APOBEC3 deaminase editing in mpox virus as evidence for sustained human transmission since at least 2016
- PMID: 37917680
- PMCID: PMC10880385
- DOI: 10.1126/science.adg8116
APOBEC3 deaminase editing in mpox virus as evidence for sustained human transmission since at least 2016
Erratum in
-
Erratum for the Research Article "APOBEC3 deaminase editing in mpox virus as evidence for sustained human transmission since at least 2016" by Á. O'Toole et al.Science. 2024 Dec 6;386(6726):eadu7667. doi: 10.1126/science.adu7667. Epub 2024 Dec 5. Science. 2024. PMID: 39637008 No abstract available.
Abstract
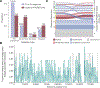
Historically, mpox has been characterized as an endemic zoonotic disease that transmits through contact with the reservoir rodent host in West and Central Africa. However, in May 2022, human cases of mpox were detected spreading internationally beyond countries with known endemic reservoirs. When the first cases from 2022 were sequenced, they shared 42 nucleotide differences from the closest mpox virus (MPXV) previously sampled. Nearly all these mutations are characteristic of the action of APOBEC3 deaminases, host enzymes with antiviral function. Assuming APOBEC3 editing is characteristic of human MPXV infection, we developed a dual-process phylogenetic molecular clock that-inferring a rate of ~6 APOBEC3 mutations per year-estimates that MPXV has been circulating in humans since 2016. These observations of sustained MPXV transmission present a fundamental shift to the perceived paradigm of MPXV epidemiology as a zoonosis and highlight the need for revising public health messaging around MPXV as well as outbreak management and control.
Conflict of interest statement
Figures


References
-
- NCDC, “An Update of Monkeypox Outbreak in Nigeria” (Nigeria Centre for Disease Control, 2017), (available at https://ncdc.gov.ng/diseases/sitreps/?cat=8).
-
- Rimoin AW, Mulembakani PM, Johnston SC, Lloyd Smith JO, Kisalu NK, Kinkela TL, Blumberg S, Thomassen HA, Pike BL, Fair JN, Wolfe ND, Shongo RL, Graham BS, Formenty P, Okitolonda E, Hensley LE, Meyer H, Wright LL, Muyembe J-J, Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. U. S. A. 107, 16262–16267 (2010). - PMC - PubMed
-
- Baker TD, “W.H.O., The Global Eradication of Smallpox. Final Report of The Global Commission for the Certification of Smallpox Eradication. Geneva, World Health Organization, 1980, 122 pp., Sfr. 11,-” in Clio Medica. Acta Academiae Internationalis Historiae Medicinae, Vol. 17 (Brill, 1982), pp. 268–269.
-
- Jezek Z, Nakano JH, Arita I, Mutombo M, Szczeniowski M, Dunn C, Serological survey for human monkeypox infections in a selected population in Zaire. J. Trop. Med. Hyg. 90, 31–38 (1987). - PubMed

