Blood-based Transcriptomic and Proteomic Biomarkers of Emphysema
- PMID: 37917913
- PMCID: PMC10840768
- DOI: 10.1164/rccm.202301-0067OC
Blood-based Transcriptomic and Proteomic Biomarkers of Emphysema
Abstract
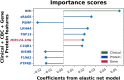
Rationale: Emphysema is a chronic obstructive pulmonary disease phenotype with important prognostic implications. Identifying blood-based biomarkers of emphysema will facilitate early diagnosis and development of targeted therapies. Objectives: To discover blood omics biomarkers for chest computed tomography-quantified emphysema and develop predictive biomarker panels. Methods: Emphysema blood biomarker discovery was performed using differential gene expression, alternative splicing, and protein association analyses in a training sample of 2,370 COPDGene participants with available blood RNA sequencing, plasma proteomics, and clinical data. Internal validation was conducted in a COPDGene testing sample (n = 1,016), and external validation was done in the ECLIPSE study (n = 526). Because low body mass index (BMI) and emphysema often co-occur, we performed a mediation analysis to quantify the effect of BMI on gene and protein associations with emphysema. Elastic net models with bootstrapping were also developed in the training sample sequentially using clinical, blood cell proportions, RNA-sequencing, and proteomic biomarkers to predict quantitative emphysema. Model accuracy was assessed by the area under the receiver operating characteristic curves for subjects stratified into tertiles of emphysema severity. Measurements and Main Results: Totals of 3,829 genes, 942 isoforms, 260 exons, and 714 proteins were significantly associated with emphysema (false discovery rate, 5%) and yielded 11 biological pathways. Seventy-four percent of these genes and 62% of these proteins showed mediation by BMI. Our prediction models demonstrated reasonable predictive performance in both COPDGene and ECLIPSE. The highest-performing model used clinical, blood cell, and protein data (area under the receiver operating characteristic curve in COPDGene testing, 0.90; 95% confidence interval, 0.85-0.90). Conclusions: Blood transcriptome and proteome-wide analyses revealed key biological pathways of emphysema and enhanced the prediction of emphysema.
Trial registration: ClinicalTrials.gov NCT00292552.
Keywords: biomarkers; emphysema; prediction; proteomics; transcriptomics.
Figures
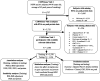
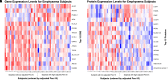
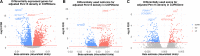
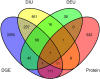

Comment in
-
Blood Biomarkers of Emphysema: What Can They Really Tell Us?Am J Respir Crit Care Med. 2024 Feb 1;209(3):235-237. doi: 10.1164/rccm.202311-2087ED. Am J Respir Crit Care Med. 2024. PMID: 38078855 Free PMC article. No abstract available.
References
-
- Lindberg A, Lindberg L, Sawalha S, Nilsson U, Stridsman C, Lundbäck B, et al. Large underreporting of COPD as cause of death—results from a population-based cohort study. Respir Med . 2021;186:106518. - PubMed
-
- Rahman HH, Niemann D, Munson-McGee SH. Association between asthma, chronic bronchitis, emphysema, chronic obstructive pulmonary disease, and lung cancer in the US population. Environ Sci Pollut Res Int . 2023;30:20147–20158. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

