Insight into the structural hierarchy of the protease cascade that regulates the mosquito melanization response
- PMID: 37918462
- PMCID: PMC10872705
- DOI: 10.1016/j.micinf.2023.105245
Insight into the structural hierarchy of the protease cascade that regulates the mosquito melanization response
Abstract
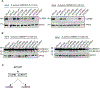
Serine protease cascades regulate important insect immune responses, including melanization and Toll pathway activation. In the context of melanization, central components of these cascades are clip domain serine proteases (CLIPs) including the catalytic, clip domain serine proteases (cSPs) and their non-catalytic homologs (cSPHs). Here, we define partially the structural hierarchy of An. gambiae cSPs of the CLIPB family, central players in melanization, and characterize their relative contributions to bacterial melanization and to mosquito susceptibility to bacterial infections. Using in vivo genetic analysis we show that the protease cascade branches downstream of the cSPs CLIPB4 and CLIPB17 into two branches one converging on CLIPB10 and the second on CLIPB8. We also show that the contribution of key cSPHs to melanization in vivo in response to diverse microbial challenges is more significant than any of the individual cSPs, possibly due to partial functional redundancy among the latter. Interestingly, we show that the key cSPH CLIPA8 which is essential for the efficient activation cleavage of CLIPBs in vivo is efficiently cleaved itself by several CLIPBs in vitro, suggesting that cSPs and cSPHs regulate signal amplification and propagation in melanization cascades by providing positive reinforcement upstream and downstream of each other.
Keywords: Anopheles gambiae; Clip domain serine protease cascades; Melanization; Mosquito innate immunity.
Copyright © 2023 Institut Pasteur. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors have no conflicts of interest to declare.
Figures
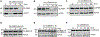
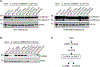



Update of
-
Insight into the structural hierarchy of the protease cascade that regulates the mosquito melanization response.bioRxiv [Preprint]. 2023 Nov 21:2023.07.13.548954. doi: 10.1101/2023.07.13.548954. bioRxiv. 2023. Update in: Microbes Infect. 2024 Jan-Feb;26(1-2):105245. doi: 10.1016/j.micinf.2023.105245. PMID: 37503117 Free PMC article. Updated. Preprint.
Similar articles
-
Insight into the structural hierarchy of the protease cascade that regulates the mosquito melanization response.bioRxiv [Preprint]. 2023 Nov 21:2023.07.13.548954. doi: 10.1101/2023.07.13.548954. bioRxiv. 2023. Update in: Microbes Infect. 2024 Jan-Feb;26(1-2):105245. doi: 10.1016/j.micinf.2023.105245. PMID: 37503117 Free PMC article. Updated. Preprint.
-
The CLIP-domain serine protease CLIPC9 regulates melanization downstream of SPCLIP1, CLIPA8, and CLIPA28 in the malaria vector Anopheles gambiae.PLoS Pathog. 2020 Oct 12;16(10):e1008985. doi: 10.1371/journal.ppat.1008985. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33045027 Free PMC article.
-
CLIPA7 Exhibits Pleiotropic Roles in the Anopheles gambiae Immune Response.J Innate Immun. 2023;15(1):317-332. doi: 10.1159/000526486. Epub 2022 Nov 24. J Innate Immun. 2023. PMID: 36423593 Free PMC article.
-
Serine proteases as mediators of mosquito immune responses.Insect Biochem Mol Biol. 2001 Mar 1;31(3):257-62. doi: 10.1016/s0965-1748(00)00145-4. Insect Biochem Mol Biol. 2001. PMID: 11167095 Review.
-
Drosophila melanogaster clip-domain serine proteases: Structure, function and regulation.Biochimie. 2016 Mar;122:255-69. doi: 10.1016/j.biochi.2015.10.007. Epub 2015 Oct 8. Biochimie. 2016. PMID: 26453810 Review.
Cited by
-
Late sporogonic stages of Plasmodium parasites are susceptible to the melanization response in Anopheles gambiae mosquitoes.bioRxiv [Preprint]. 2024 May 31:2024.05.31.596773. doi: 10.1101/2024.05.31.596773. bioRxiv. 2024. Update in: Front Cell Infect Microbiol. 2024 Aug 01;14:1438019. doi: 10.3389/fcimb.2024.1438019. PMID: 38853990 Free PMC article. Updated. Preprint.
-
CLIPB4 Is a Central Node in the Protease Network that Regulates Humoral Immunity in Anopheles gambiae Mosquitoes.J Innate Immun. 2023;15(1):680-696. doi: 10.1159/000533898. Epub 2023 Sep 13. J Innate Immun. 2023. PMID: 37703846 Free PMC article.
-
Advances in the dissection of Anopheles-Plasmodium interactions.PLoS Pathog. 2025 Mar 31;21(3):e1012965. doi: 10.1371/journal.ppat.1012965. eCollection 2025 Mar. PLoS Pathog. 2025. PMID: 40163471 Free PMC article. Review.
-
CLIPB4 is a central node in the protease network that regulates humoral immunity in Anopheles gambiae mosquitoes.bioRxiv [Preprint]. 2023 Jul 16:2023.07.07.545904. doi: 10.1101/2023.07.07.545904. bioRxiv. 2023. Update in: J Innate Immun. 2023;15(1):680-696. doi: 10.1159/000533898. PMID: 37461554 Free PMC article. Updated. Preprint.
-
Late sporogonic stages of Plasmodium parasites are susceptible to the melanization response in Anopheles gambiae mosquitoes.Front Cell Infect Microbiol. 2024 Aug 1;14:1438019. doi: 10.3389/fcimb.2024.1438019. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39149419 Free PMC article.
References
-
- Dudzic JP, Hanson MA, Iatsenko I, Kondo S, Lemaitre B. More Than Black or White: Melanization and Toll Share Regulatory Serine Proteases in Drosophila. Cell Rep 2019;27:1050–61 e3. - PubMed
-
- Veillard F, Troxler L, Reichhart JM. Drosophila melanogaster clip-domain serine proteases: Structure, function and regulation. Biochimie 2016;122:255–69. - PubMed
-
- Nakhleh J, El Moussawi L, Osta MA. The melanization response in insect immunity. in: Ligoxygakis P (Ed.), Insect Immunity, Elsevier, 2017, pp. 2–20.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

