The lncRNA Sweetheart regulates compensatory cardiac hypertrophy after myocardial injury in murine males
- PMID: 37919291
- PMCID: PMC10622434
- DOI: 10.1038/s41467-023-42760-y
The lncRNA Sweetheart regulates compensatory cardiac hypertrophy after myocardial injury in murine males
Abstract
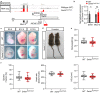
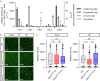
After myocardial infarction in the adult heart the remaining, non-infarcted tissue adapts to compensate the loss of functional tissue. This adaptation requires changes in gene expression networks, which are mostly controlled by transcription regulating proteins. Long non-coding transcripts (lncRNAs) are taking part in fine-tuning such gene programs. We describe and characterize the cardiomyocyte specific lncRNA Sweetheart RNA (Swhtr), an approximately 10 kb long transcript divergently expressed from the cardiac core transcription factor coding gene Nkx2-5. We show that Swhtr is dispensable for normal heart development and function but becomes essential for the tissue adaptation process after myocardial infarction in murine males. Re-expressing Swhtr from an exogenous locus rescues the Swhtr null phenotype. Genes that depend on Swhtr after cardiac stress are significantly occupied and therefore most likely regulated by NKX2-5. The Swhtr transcript interacts with NKX2-5 and disperses upon hypoxic stress in cardiomyocytes, indicating an auxiliary role of Swhtr for NKX2-5 function in tissue adaptation after myocardial injury.
© 2023. The Author(s).
Conflict of interest statement
The authors declare the following financial interest; S.O. is currently the CEO and shareholder of HAYA Therapeutics. HAYA is developing ASO-based drugs to target lncRNAs in heart disease. S.O. declares non-financial competing interests. The other authors declare no competing interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

