DNA-encoded library-enabled discovery of proximity-inducing small molecules
- PMID: 37919549
- PMCID: PMC10917151
- DOI: 10.1038/s41589-023-01458-4
DNA-encoded library-enabled discovery of proximity-inducing small molecules
Abstract
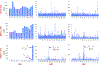
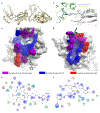
Small molecules that induce protein-protein associations represent powerful tools to modulate cell circuitry. We sought to develop a platform for the direct discovery of compounds able to induce association of any two preselected proteins, using the E3 ligase von Hippel-Lindau (VHL) and bromodomains as test systems. Leveraging the screening power of DNA-encoded libraries (DELs), we synthesized ~1 million DNA-encoded compounds that possess a VHL-targeting ligand, a variety of connectors and a diversity element generated by split-and-pool combinatorial chemistry. By screening our DEL against bromodomains in the presence and absence of VHL, we could identify VHL-bound molecules that simultaneously bind bromodomains. For highly barcode-enriched library members, ternary complex formation leading to bromodomain degradation was confirmed in cells. Furthermore, a ternary complex crystal structure was obtained for our most enriched library member with BRD4BD1 and a VHL complex. Our work provides a foundation for adapting DEL screening to the discovery of proximity-inducing small molecules.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare the following competing financial interests: X.M. is a shareholder of Terremoto Biosciences. C.W.C. is an advisor to Anagenex. P.A.C. is an advisor to nference, Inc., Pfizer, Inc., and Belharra Therapeutics. S.L.S. is a shareholder and serves on the Board of Directors of Jnana Therapeutics and Kojin Therapeutics; is a shareholder and advises Kisbee Therapeutics, Belharra Therapeutics, Magnet Biomedicine, Exo Therapeutics, and Eikonizo Therapeutics; advises Vividian Therapeutics, Eisai Co., Ltd., Ono Pharma Foundation, F-Prime Capital Partners, and the Genomics Institute of the Novartis Research Foundation; and is a Novartis Faculty Scholar. The remaining authors declare no competing interests.
Figures


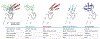





References
-
- Schreiber SL The Rise of Molecular Glues. Cell 184, 3–9 (2021). - PubMed
-
- Liu J et al. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66, 807–815 (1991). - PubMed
-
- Brown EJ et al. A mammalian protein targeted by G1-arresting rapamycin–receptor complex. Nature 369, 756–758 (1994). - PubMed
-
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P & Snyder SH RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78, 35–43 (1994). - PubMed
-
- Haggarty SJ et al. Dissecting cellular processes using small molecules: identification of colchicine-like, taxol-like and other small molecules that perturb mitosis. Chemistry & Biology 7, 275–286 (2000). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

