MYC overexpression and SMARCA4 loss cooperate to drive medulloblastoma formation in mice
- PMID: 37919824
- PMCID: PMC10621315
- DOI: 10.1186/s40478-023-01654-2
MYC overexpression and SMARCA4 loss cooperate to drive medulloblastoma formation in mice
Abstract
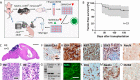
Group 3 medulloblastoma is one of the most aggressive types of childhood brain tumors. Roughly 30% of cases carry genetic alterations in MYC, SMARCA4, or both genes combined. While overexpression of MYC has previously been shown to drive medulloblastoma formation in mice, the functional significance of SMARCA4 mutations and their suitability as a therapeutic target remain largely unclear. To address this issue, we combined overexpression of MYC with a loss of SMARCA4 in granule cell precursors. Both alterations did not increase proliferation of granule cell precursors in vitro. However, combined MYC overexpression and SMARCA4 loss successfully induced tumor formation in vivo after orthotopic transplantation in recipient mice. Resulting tumors displayed anaplastic histology and exclusively consisted of SMARCA4-negative cells although a mixture of recombined and non-recombined cells was injected. These observations provide first evidence for a tumor-promoting role of a SMARCA4 deficiency in the development of medulloblastoma. In comparing the transcriptome of tumors to the cells of origin and an established Sonic Hedgehog medulloblastoma model, we gathered first hints on deregulated gene expression that could be specifically involved in SMARCA4/MYC driven tumorigenesis. Finally, an integration of RNA sequencing and DNA methylation data of murine tumors with human samples revealed a high resemblance to human Group 3 medulloblastoma on the molecular level. Altogether, the development of SMARCA4-deficient medulloblastomas in mice paves the way to deciphering the role of frequently occurring SMARCA4 alterations in Group 3 medulloblastoma with the perspective to explore targeted therapeutic options.
Keywords: BAF complex; BRG1; Chromatin remodeling; Group 3 medulloblastoma; MYC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

