Conservation of knotted and slipknotted topology in transmembrane transporters
- PMID: 37919904
- PMCID: PMC10719070
- DOI: 10.1016/j.bpj.2023.10.031
Conservation of knotted and slipknotted topology in transmembrane transporters
Abstract
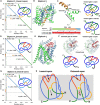
The existence of nontrivial topology is well accepted in globular proteins but not in membrane proteins. Our comprehensive topological analysis of the Protein Data Bank structures reveals 18 families of transmembrane proteins with nontrivial topology, showing that they constitute a significant number of membrane proteins. Moreover, we found that they comprise one of the largest groups of secondary active transporters. We classified them based on their knotted fingerprint into four groups: three slipknotted and one knotted. Unexpectedly, we found that the same protein can possess two distinct slipknot motifs that correspond to its outward- and inward-open conformational state. Based on the analysis of structures and knotted fingerprints, we show that slipknot topology is directly involved in the conformational transition and substrate transfer. Therefore, entanglement can be used to classify proteins and to find their structure-function relationship. Furthermore, based on the topological analysis of the transmembrane protein structures predicted by AlphaFold, we identified new potentially slipknotted protein families.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

