The purinergic receptor P2X7 and the NLRP3 inflammasome are druggable host factors required for SARS-CoV-2 infection
- PMID: 37920468
- PMCID: PMC10619763
- DOI: 10.3389/fimmu.2023.1270081
The purinergic receptor P2X7 and the NLRP3 inflammasome are druggable host factors required for SARS-CoV-2 infection
Abstract
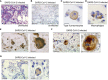
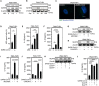
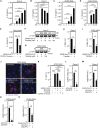
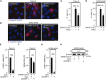
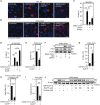
Purinergic receptors and NOD-like receptor protein 3 (NLRP3) inflammasome regulate inflammation and viral infection, but their effects on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection remain poorly understood. Here, we report that the purinergic receptor P2X7 and NLRP3 inflammasome are cellular host factors required for SARS-CoV-2 infection. Lung autopsies from patients with severe coronavirus disease 2019 (COVID-19) reveal that NLRP3 expression is increased in host cellular targets of SARS-CoV-2 including alveolar macrophages, type II pneumocytes and syncytia arising from the fusion of infected macrophages, thus suggesting a potential role of NLRP3 and associated signaling pathways to both inflammation and viral replication. In vitro studies demonstrate that NLRP3-dependent inflammasome activation is detected upon macrophage abortive infection. More importantly, a weak activation of NLRP3 inflammasome is also detected during the early steps of SARS-CoV-2 infection of epithelial cells and promotes the viral replication in these cells. Interestingly, the purinergic receptor P2X7, which is known to control NLRP3 inflammasome activation, also favors the replication of D614G and alpha SARS-CoV-2 variants. Altogether, our results reveal an unexpected relationship between the purinergic receptor P2X7, the NLRP3 inflammasome and the permissiveness to SARS-CoV-2 infection that offers novel opportunities for COVID-19 treatment.
Keywords: COVID-19; NLRP3; P2X7; SARS-CoV-2; inflammasome.
Copyright © 2023 Lécuyer, Nardacci, Tannous, Gutierrez-Mateyron, Deva Nathan, Subra, Di Primio, Quaranta, Petit, Richetta, Mostefa-Kara, Del Nonno, Falasca, Marlin, Maisonnasse, Delahousse, Pascaud, Deprez, Naigeon, Chaput, Paci, Saada, Ghez, Mariette, Costa, Pistello, Allouch, Delelis, Piacentini, Le Grand and Perfettini.
Conflict of interest statement
DT and AA were employed by NH TherAguix SAS. Authors DL, DT, AA, FS, OD and J-LP are listed as co-inventors on a patent application related to SARS-CoV-2 therapy. AP and J-LP are founding members of Findimmune SAS, an Immuno-Oncology Biotech company. J-LP disclosed research funding not related to this work from NH TherAguix and Wonna Therapeutics. NC disclosed research funding not related to this work from GlaxoSmithKline, Roche, Cytune pharma and Sanofi. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

