Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation
- PMID: 37921038
- PMCID: PMC10702828
- DOI: 10.15252/embr.202357984
Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation
Abstract
The rixosome defined in Schizosaccharomyces pombe and humans performs diverse roles in pre-ribosomal RNA processing and gene silencing. Here, we isolate and describe the conserved rixosome from Chaetomium thermophilum, which consists of two sub-modules, the sphere-like Rix1-Ipi3-Ipi1 and the butterfly-like Las1-Grc3 complex, connected by a flexible linker. The Rix1 complex of the rixosome utilizes Sda1 as landing platform on nucleoplasmic pre-60S particles to wedge between the 5S rRNA tip and L1-stalk, thereby facilitating the 180° rotation of the immature 5S RNP towards its mature conformation. Upon rixosome positioning, the other sub-module with Las1 endonuclease and Grc3 polynucleotide-kinase can reach a strategic position at the pre-60S foot to cleave and 5' phosphorylate the nearby ITS2 pre-rRNA. Finally, inward movement of the L1 stalk permits the flexible Nop53 N-terminus with its AIM motif to become positioned at the base of the L1-stalk to facilitate Mtr4 helicase-exosome participation for completing ITS2 removal. Thus, the rixosome structure elucidates the coordination of two central ribosome biogenesis events, but its role in gene silencing may adapt similar strategies.
Keywords: 5S RNP; ITS2; pre-60S ribosome; ribosome biogenesis; rixosome.
© 2023 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Affinity‐purification of wild‐type PTF‐Rsa4 (lane 1) and the PTF‐Rsa4 E117D mutant from C. thermophilum lysates (lane 2 and 3; two different transformants). The final eluates were analyzed by SDS–PAGE and Coomassie staining. The labeled bands were identified by mass spectrometry. In the case of wild‐type PTF‐Rsa4, Scp160 (band 1), Hsp90 (band 2), and Hsp70 (band 3) were identified by mass spectrometry as possible contaminants. The PTF‐Rsa4 wt purifies predominantly mature ribosomes whereas the PTF‐Rsa4 E117D mainly purifies pre‐60S particles.
The split‐pair combination PT‐Rsa4 E117D and Nop7‐Flag was used to affinity‐purify pre‐60S particles from C. thermophilum lysates. Both the first TEV eluate (lane 1) and second Flag eluate (lane 2) were analyzed by SDS–PAGE and Coomassie staining. Bands labeled on the right were identified by mass spectrometry, with bait proteins labeled in blue and Las1‐Grc3 complex in red, respectively. The affinity‐purifications have been performed at least twice with similar outcome.
Growth analysis of ectopically expressed RSA4 wild‐type and rsa4 E117D mutant genes in C. thermophilum. Small mycelium pieces of similar size, containing the indicated pre‐grown C. thermophilum strains, were plated on CCM agar and mycelium growth was analyzed after 13 h at 50°C.

Split‐tag affinity‐purification using PT‐Rsa4 E117D as first bait, followed by Rix1‐Flag (left) or Flag‐Las1 (right) as second bait enriching pre‐60S particles that contain the Rix1‐Ipi1‐Ipi3‐Las1‐Grc3 (rixosome) complex. Both the first TEV and second Flag eluate were analyzed by SDS–PAGE and Coomassie staining. Labeled bands were identified by mass spectrometry, with bait proteins labeled in blue. Both affinity‐purifications were performed at least twice with similar outcome.
Sucrose gradient centrifugation (10–40%) of the final PT‐Rsa4 E117D Rix1‐Flag split‐tag eluate as shown in (A). The input and gradient fractions 1–15 were analyzed by SDS–PAGE and Coomassie staining. The labeled bands were excised from the gel and identified by mass spectrometry.
Affinity‐purification of PTF‐Las1 from wild‐type C. thermophilum lysates (final eluate, Input), which was further fractionated by sucrose gradient centrifugation (10–40%). Input and gradient fractions 1–15 were finally analyzed by SDS–PAGE and Coomassie staining. Bands labeled on the left were identified by mass spectrometry.
Negative‐stain electron microscopy of sucrose gradient fraction #4 (see C) from the final PTF‐Las1 Flag eluate containing the highly purified rixosome complex (see also C). Shown are typical 2D classes of the rixosome, consisting of the Rix1 subcomplex (hollow sphere) in association with the Las1‐Grc3 heterodimer (butterfly‐like). Scale bar, 50 nm.

- A
Selected 2D classes of the rixosome complex.
- B
Low‐resolution cryo‐EM map of the rixosome. The surface is colored according to the Las1‐Grc3 complex shown in (D). The approximate distance between the Las1‐Grc3 complex and the globular Rix1 complex is indicated, and the unresolved Las1 CC domain is indicated as dashed line.
- C, D
Colored cryo‐EM maps (left) and molecular models (right) of the individually processed Rix1 complex (C) and the Las1‐Grc3 complex (D) shown in two orientations.
- E
The Las1‐Grc3 was found in the apo‐state. Close‐up of the nucleotide‐binding pocket of Grc3 (middle) and overlay with the ATP‐γS model of PDB‐ID 6OF2 (bottom).

Schematic representation and molecular models of Grc3, Las1, Ipi3, and Rix1 shown in different colors. Regions not included in the molecular models are indicated in gray.
Alphafold2‐multimer model of the Las1 coiled‐coil domain (Las1‐CC, aa179‐343) in complex with a dimer of the C‐terminal coiled‐coil helix of Ipi3 (Ipi3‐CC, aa394‐437). The maximal length of the unstructured regions are indicated in Å.
Alphafold2‐multimer predictions of the Las1 C‐term/Ipi3‐CC interactions from S. pombe, H. sapiens, and S. cerevisiae.

- A–C
Cryo‐EM maps of three sequential pre‐60 states shown in two orientations. The maps are colored according to the respective molecular models and biogenesis factors, 5S rRNA and ITS2 are labeled. Ribosomal rRNA and ribosomal proteins are shown in gray.
- D
Model illustrating the molecular steps in the rotation of the 5S rRNP throughout the three states. The 5S rRNA, Rpf2‐Rrs1, Rix1 complex, and Sda1 are shown.

Nucleoplasmic pre‐60S particle from C. thermophilum lack the polypeptide exit tunnel (PET) bound export factor Arx1. Comparison between nucleoplasmic pre‐60S states from ct (left, state 1) and yeast (middle, PDB‐ID: 7UOO) and cytoplasmic pre‐60S particles after release of Nog1 and Rlp24 from human bound to EBP1 (human Arx1 homolog, right, PDB‐ID: 6LSR). The upper panels show the molecular models of the different states, and the lower panels focus on the polypeptide exit tunnel.
Superposition of Arx1‐Alb1 model from yeast and the Ebp1 model from human onto the pre‐5S rotation state from C. thermophilum (state 1). The Nog1 C‐terminus from C. thermophilum sterically clashes with the binding sites of Arx1 and EBP1 rationalizing the lack of ct Arx1 on the here described nucleoplasmic pre‐60S states.
Comparison of the ct ITS2 containing Foot structure (state 1) and the equivalent yeast state (Nog2pre, PDB‐ID: 7UOO).
Colored cryo‐EM map of the ct pre‐5S rotation state 1 in two orientations highlighting the two copies of uL30 within the particle (upper panels). Detailed view of the individual uL30 copies (lower panel, left and middle) and merge of the two copies (lower panel, right). The N‐terminus of uL30Foot has to rotate in comparison to the uL30 copy within the pre‐60S core to adapt the binding mode of Rlp7 within the yeast Foot.
Structural comparison of ct Utp30 bound to helix 2 (H2) of the ITS2 within the Foot structure, ct Utp30 as part of the ct 90S bound to es10 (PDB‐ID: 6RXU) and the yeast foot factor Nsa3 bound to the H2 of the ITS2 (PDB‐ID: 7UOO). The models are labeled and shown in two orientations and as merge comparing pre‐60S ct Utp30/ITS2‐H2 with 90S ct Utp30/es10 and pre‐60S sc Nsa3/ITS2‐H2, respectively.

Molecular model of the Rix1 complex on the pre‐5S and post‐5S rotation particles. Close‐up showing the Ipi1 loop (aa241‐278) interacting with the Rix1‐Ipi3 sphere.
Colored maps of the pre‐5S and post‐5S rotation particles highlighting the Rix1 complex and molecular models of the Rix1 complex from both states illustrating its movement.
Sda1 and the L1 stalk rearrange during 5S rRNA rotation (left and middle panel). The pre‐60S moieties of both states are shown as gray density and 5S rRNA, Rpf2‐Rrs1, and Nog2 are colored. Sda1‐Ipi1 and the L1 stalk are shown as molecular model and the Rix1‐Ipi3 complex was omitted for clarity. Overlay of the Sda1‐Ipi1 complex from the pre‐5S and post‐5S rotation particles (right panel). The two Ipi1's were rigid body fitted to indicate the movement of Sda1 between both states.
The Rix1 complex interacts with the 5S rRNA tip and Rpf2‐Rrs1 in the pre‐5S rRNA rotation state (left side). Models of 5S rRNA, Rpf2‐Rrs1, and Rix1‐Ipi3 and the Gaussian filtered segmented densities of 5S rRNA and Rpf2‐Rrs1 are shown in two orientations. One copy of each Ipi3 and Rix1 form a positively charged surface interacting with the 5S rRNA tip (right side). Magnifications show the surface views of Rix1 and Ipi3 colored according to the respective electrostatic potential.
The positively charged N‐terminal half of one of the Rix1 copies is in contact with the L1 stalk rRNA in the pre‐5S and post‐5S rotation state. The surface colored according to the electrostatic potential for the interacting Rix1 copy and transparent models for the other Rix1 complex members are shown. The L1 stalk is shown as molecular model.
Overview of the intrinsic 5S rRNP rotation network comparing the pre‐5S (right side) and post‐5S rotation state (left side). Factors involved in stabilization and rotation of the 5S RNP are shown as colored molecular models and the pre‐60S moieties as filtered transparent densities.
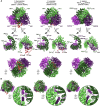
The Rix1 complex shows strong structural conservation between organisms. Comparison of the molecular models of the Rix1 complex from C. thermophilum, S. cerevisiae, and the PELP1‐WDR18 complex from H. sapiens.
Detailed view of the C‐termini of Ipi3/WDR18 for each species. The termini are similarly positioned and able to protrude from the Rix1 complex, potentially form coiled‐coils and interact with Las1 (see Fig EV1).

Superposition of the rixosome density onto the colored map of the pre‐5S rotation state with an engaged Rix1 complex shown in two orientations. The globular ball‐like part of the rixosome map was superimposed with the Rix1 complex density and the butterfly‐like Las1‐Grc3 density faces toward the ITS2‐foot structure. The colored model of the Las1‐Grc3 complex was rigid body fitted.
Close‐up of the Rix1 complex area within the pre‐5S rotation state, showing the molecular model and the corresponding density low pass filtered to 25 Å. An asterisk denotes an additional density protruding from the Rix1 complex at the location where the C‐terminal coiled‐coil helices are presumed to emerge.
Low pass filtered and colored maps of different pre‐60S states. The movement of the L1 stalk is indicated. The N‐terminus of Nop53 (modeled from aa3‐52) binds the pre‐60S particle when the L1 stalk is in an inward position either contacting the 5S rRNA tip in the pre‐5S rotation state (middle) or in the post‐5S rotation state (right), marked with an asterisk.
Colored surface model of the pre‐60S state with the L1 stalk in the inward position. The molecular model on Nop53 is shown. Close‐up views highlight the N‐terminus of Nop53 and its interaction partners. The unresolved region between aa52 and aa85 containing the AIM motif is indicated as dashed line. The N‐ and C‐termini of the Nop53 model are labeled as N3 and C433, respectively.

- A–E
Colored cryo‐EM maps of additional pre‐60S states identified in the study. (A) Close‐ups showing the colored density map of the partly processed foot structure of the pre‐5S rotation‐lacking Utp30/ITS2 state. Superimposed models of the pre‐5S rotation state (Arx1/Nog2 state) indicating the missing parts: Utp30, ITS2, the C‐terminal part of Nop15 (aa277‐335), and Nop53 (aa85‐107). (D) Close‐up of the immature H68/H69 area with superimposed transparent model of H68 and H69 of the pre‐5S rotation state (Arx1/Nog2 state).

Colored surface representation of the pre‐5S rotation state with L1 in the inward position (State 5). Nop53 is shown as model. The superimposed KOW domain together with the Nop53 AIM motif from S. cerevisiae is shown (PDB‐ID: 5OOQ). Only aa666‐818 Mtr4‐KOW and aa59‐70 Nop53‐AIM are shown for clarity. The model was rigid body fitted into the merged pre‐60S‐Exo‐14n structure. Dashed lines indicate the flexible, unmodeled parts of Nop53 wrapping around the Mtr4‐KOW domain.
Filtered composite map of the pre‐60S‐Exo‐14n complex from S. cerevisiae arrested at the 5.8S+30 pre‐rRNA step (EMDB‐IDs: 4301 and 4302, PDB‐IDs: 6FT6 and 6FSZ).
The filtered pre‐60S‐Exo‐14n complex composite map superimposed with the C. thermophilum pre‐5S rotation–L1 inward state (State 5) shown in dark gray and the L1 stalk, 5S rRNA, and Rsa4 in brown, yellow, and green, respectively.
Overview (upper panel) and detailed view (lower panel) of the merge between the pre‐60S‐Exo‐14n density and the pre‐5S rotation state with engaged Rix1 complex (State 3). Only the Rix1 complex model is shown for clarity. Mtr4 and the Rix1 complex do not sterically interfere with each other.
References
-
- Al‐Samarrai TH, Schmid J (2000) A simple method for extraction of fungal genomic DNA. Lett Appl Microbiol 30: 53–56 - PubMed
-
- Barrio‐Garcia C, Thoms M, Flemming D, Kater L, Berninghausen O, Bassler J, Beckmann R, Hurt E (2016) Architecture of the Rix1‐Rea1 checkpoint machinery during pre‐60S‐ribosome remodeling. Nat Struct Mol Biol 23: 37–44 - PubMed
-
- Bassler J, Hurt E (2019) Eukaryotic ribosome assembly. Annu Rev Biochem 88: 281–306 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

