The antiproliferative effect of FGF2 in K-Ras-driven tumor cells involves modulation of rRNA and the nucleolus
- PMID: 37921359
- PMCID: PMC11166202
- DOI: 10.1242/jcs.260989
The antiproliferative effect of FGF2 in K-Ras-driven tumor cells involves modulation of rRNA and the nucleolus
Abstract
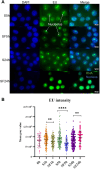
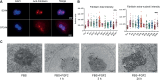
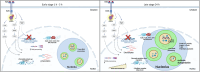
The nucleolus is sensitive to stress and can orchestrate a chain of cellular events in response to stress signals. Despite being a growth factor, FGF2 has antiproliferative and tumor-suppressive functions in some cellular contexts. In this work, we investigated how the antiproliferative effect of FGF2 modulates chromatin-, nucleolus- and rDNA-associated proteins. The chromatin and nucleolar proteome indicated that FGF2 stimulation modulates proteins related to transcription, rRNA expression and chromatin-remodeling proteins. The global transcriptional rate and nucleolus area increased along with nucleolar disorganization upon 24 h of FGF2 stimulation. FGF2 stimulation induced immature rRNA accumulation by increasing rRNA transcription. The rDNA-associated protein analysis reinforced that FGF2 stimulus interferes with transcription and rRNA processing. RNA Pol I inhibition partially reversed the growth arrest induced by FGF2, indicating that changes in rRNA expression might be crucial for triggering the antiproliferative effect. Taken together, we demonstrate that the antiproliferative FGF2 stimulus triggers significant transcriptional changes and modulates the main cell transcription site, the nucleolus.
Keywords: Chromatin; FGF2; Nucleolus; Proteomics; Transcription.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




Similar articles
-
Transcriptional suppression of ribosomal DNA with phase separation.Sci Adv. 2020 Oct 14;6(42):eabb5953. doi: 10.1126/sciadv.abb5953. Print 2020 Oct. Sci Adv. 2020. PMID: 33055158 Free PMC article.
-
Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders.Nature. 2018 Feb 1;554(7690):112-117. doi: 10.1038/nature25449. Epub 2018 Jan 24. Nature. 2018. PMID: 29364875 Free PMC article.
-
FGF2 Antiproliferative Stimulation Induces Proteomic Dynamic Changes and High Expression of FOSB and JUNB in K-Ras-Driven Mouse Tumor Cells.Proteomics. 2018 Sep;18(17):e1800203. doi: 10.1002/pmic.201800203. Epub 2018 Aug 14. Proteomics. 2018. PMID: 30035358
-
Chromatin: linking structure and function in the nucleolus.Chromosoma. 2009 Feb;118(1):11-23. doi: 10.1007/s00412-008-0184-2. Epub 2008 Oct 17. Chromosoma. 2009. PMID: 18925405 Review.
-
The nucleolus—guardian of cellular homeostasis and genome integrity.Chromosoma. 2013 Dec;122(6):487-97. doi: 10.1007/s00412-013-0430-0. Chromosoma. 2013. PMID: 24022641 Review.
Cited by
-
Regulation of Hair Follicle Growth and Development by Different Alternative Spliceosomes of FGF5 in Rabbits.Genes (Basel). 2024 Mar 26;15(4):409. doi: 10.3390/genes15040409. Genes (Basel). 2024. PMID: 38674344 Free PMC article.
References
-
- Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., Fridman, W.-H., Pagès, F., Trajanoski, Z. and Galon, J. (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics (Oxford, England) 25, 1091-1093. 10.1093/bioinformatics/btp101 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

