Genomic stability of self-inactivating rabies
- PMID: 37921437
- PMCID: PMC10666929
- DOI: 10.7554/eLife.83459
Genomic stability of self-inactivating rabies
Abstract
Transsynaptic viral vectors provide means to gain genetic access to neurons based on synaptic connectivity and are essential tools for the dissection of neural circuit function. Among them, the retrograde monosynaptic ΔG-Rabies has been widely used in neuroscience research. A recently developed engineered version of the ΔG-Rabies, the non-toxic self-inactivating (SiR) virus, allows the long term genetic manipulation of neural circuits. However, the high mutational rate of the rabies virus poses a risk that mutations targeting the key genetic regulatory element in the SiR genome could emerge and revert it to a canonical ΔG-Rabies. Such revertant mutations have recently been identified in a SiR batch. To address the origin, incidence and relevance of these mutations, we investigated the genomic stability of SiR in vitro and in vivo. We found that "revertant" mutations are rare and accumulate only when SiR is extensively amplified in vitro, particularly in suboptimal production cell lines that have insufficient levels of TEV protease activity. Moreover, we confirmed that SiR-CRE, unlike canonical ΔG-Rab-CRE or revertant-SiR-CRE, is non-toxic and that revertant mutations do not emerge in vivo during long-term experiments.
Keywords: genetics; genomics; mouse; neural circuits; neuronal tracing; neuroscience; rabies; viral vector.
© 2023, Ciabatti et al.
Conflict of interest statement
EC The SiR technology is patented by the UK Research and Innovation (WO2018203049A1), AG, Dd, HL, FM, MT No competing interests declared
Figures




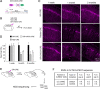
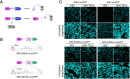

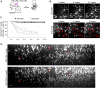

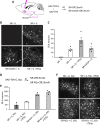
Update of
References
-
- Abdelfattah AS, Kawashima T, Singh A, Novak O, Liu H, Shuai Y, Huang Y-C, Campagnola L, Seeman SC, Yu J, Zheng J, Grimm JB, Patel R, Friedrich J, Mensh BD, Paninski L, Macklin JJ, Murphy GJ, Podgorski K, Lin B-J, Chen T-W, Turner GC, Liu Z, Koyama M, Svoboda K, Ahrens MB, Lavis LD, Schreiter ER. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science. 2019;365:699–704. doi: 10.1126/science.aav6416. - DOI - PubMed
-
- Andersen Laboratory Ivar. 08aac33GitHub. 2023 https://github.com/andersen-lab/ivar
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

