Neutron crystallographic refinement with REFMAC5 from the CCP4 suite
- PMID: 37921806
- PMCID: PMC7615533
- DOI: 10.1107/S2059798323008793
Neutron crystallographic refinement with REFMAC5 from the CCP4 suite
Abstract
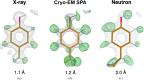
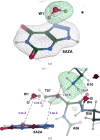
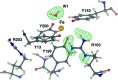
Hydrogen (H) atoms are abundant in macromolecules and often play critical roles in enzyme catalysis, ligand-recognition processes and protein-protein interactions. However, their direct visualization by diffraction techniques is challenging. Macromolecular X-ray crystallography affords the localization of only the most ordered H atoms at (sub-)atomic resolution (around 1.2 Å or higher). However, many H atoms of biochemical significance remain undetectable by this method. In contrast, neutron diffraction methods enable the visualization of most H atoms, typically in the form of deuterium (2H) atoms, at much more common resolution values (better than 2.5 Å). Thus, neutron crystallography, although technically demanding, is often the method of choice when direct information on protonation states is sought. REFMAC5 from the Collaborative Computational Project No. 4 (CCP4) is a program for the refinement of macromolecular models against X-ray crystallographic and cryo-EM data. This contribution describes its extension to include the refinement of structural models obtained from neutron crystallographic data. Stereochemical restraints with accurate bond distances between H atoms and their parent atom nuclei are now part of the CCP4 Monomer Library, the source of prior chemical information used in the refinement. One new feature for neutron data analysis in REFMAC5 is refinement of the protium/deuterium (1H/2H) fraction. This parameter describes the relative 1H/2H contribution to neutron scattering for hydrogen isotopes. The newly developed REFMAC5 algorithms were tested by performing the (re-)refinement of several entries available in the PDB and of one novel structure (FutA) using either (i) neutron data only or (ii) neutron data supplemented by external restraints to a reference X-ray crystallographic structure. Re-refinement with REFMAC5 afforded models characterized by R-factor values that are consistent with, and in some cases better than, the originally deposited values. The use of external reference structure restraints during refinement has been observed to be a valuable strategy, especially for structures at medium-low resolution.
Keywords: CCP4; H atoms; REFMAC5; crystallographic refinement; neutron macromolecular crystallography.
open access.
Figures

References
-
- Agirre, J., Atanasova, M., Bagdonas, H., Ballard, C. B., Baslé, A., Beilsten-Edmands, J., Borges, R. J., Brown, D. G., Burgos-Mármol, J. J., Berrisford, J. M., Bond, P. S., Caballero, I., Catapano, L., Chojnowski, G., Cook, A. G., Cowtan, K. D., Croll, T. I., Debreczeni, J. É., Devenish, N. E., Dodson, E. J., Drevon, T. R., Emsley, P., Evans, G., Evans, P. R., Fando, M., Foadi, J., Fuentes-Montero, L., Garman, E. F., Gerstel, M., Gildea, R. J., Hatti, K., Hekkelman, M. L., Heuser, P., Hoh, S. W., Hough, M. A., Jenkins, H. T., Jiménez, E., Joosten, R. P., Keegan, R. M., Keep, N., Krissinel, E. B., Kolenko, P., Kovalevskiy, O., Lamzin, V. S., Lawson, D. M., Lebedev, A. A., Leslie, A. G. W., Lohkamp, B., Long, F., Malý, M., McCoy, A. J., McNicholas, S. J., Medina, A., Millán, C., Murray, J. W., Murshudov, G. N., Nicholls, R. A., Noble, M. E. M., Oeffner, R., Pannu, N. S., Parkhurst, J. M., Pearce, N., Pereira, J., Perrakis, A., Powell, H. R., Read, R. J., Rigden, D. J., Rochira, W., Sammito, M., Sánchez Rodríguez, F., Sheldrick, G. M., Shelley, K. L., Simkovic, F., Simpkin, A. J., Skubak, P., Sobolev, E., Steiner, R. A., Stevenson, K., Tews, I., Thomas, J. M. H., Thorn, A., Valls, J. T., Uski, V., Usón, I., Vagin, A., Velankar, S., Vollmar, M., Walden, H., Waterman, D., Wilson, K. S., Winn, M. D., Winter, G., Wojdyr, M. & Yamashita, K. (2023). Acta Cryst. D79, 449–461.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

