POLE3 is a repressor of unintegrated HIV-1 DNA required for efficient virus integration and escape from innate immune sensing
- PMID: 37922361
- PMCID: PMC10624344
- DOI: 10.1126/sciadv.adh3642
POLE3 is a repressor of unintegrated HIV-1 DNA required for efficient virus integration and escape from innate immune sensing
Abstract
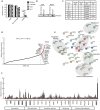
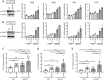
Unintegrated retroviral DNA is transcriptionally silenced by host chromatin silencing factors. Here, we used the proteomics of isolated chromatin segments method to reveal viral and host factors associated with unintegrated HIV-1DNA involved in its silencing. By gene silencing using siRNAs, 46 factors were identified as potential repressors of unintegrated HIV-1DNA. Knockdown and knockout experiments revealed POLE3 as a transcriptional repressor of unintegrated HIV-1DNA. POLE3 maintains unintegrated HIV-1DNA in a repressive chromatin state, preventing RNAPII recruitment to the viral promoter. POLE3 and the recently identified host factors mediating unintegrated HIV-1 DNA silencing, CAF1 and SMC5/SMC6/SLF2, show specificity toward different forms of unintegrated HIV-1DNA. Loss of POLE3 impaired HIV-1 replication, suggesting that repression of unintegrated HIV-1DNA is important for optimal viral replication. POLE3 depletion reduces the integration efficiency of HIV-1. POLE3, by maintaining a repressive chromatin structure of unintegrated HIV-1DNA, ensures HIV-1 escape from innate immune sensing in primary CD4+ T cells.
Figures




References
-
- M. Lusic, R. F. Siliciano, Nuclear landscape of HIV-1 infection and integration. Nat. Rev. Microbiol. 15, 69–82 (2017). - PubMed
-
- S. Machida, D. Depierre, H.-C. Chen, S. Thenin-Houssier, G. Petitjean, C. M. Doyen, M. Takaku, O. Cuvier, M. Benkirane, Exploring histone loading on HIV DNA reveals a dynamic nucleosome positioning between unintegrated and integrated viral genome. Proc. Natl. Acad. Sci. U.S.A. 117, 6822–6830 (2020). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

