The role of midbody-associated mRNAs in regulating abscission
- PMID: 37922419
- PMCID: PMC10624257
- DOI: 10.1083/jcb.202306123
The role of midbody-associated mRNAs in regulating abscission
Abstract
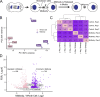
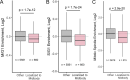
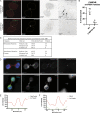
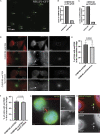
Midbodies function during telophase to regulate the abscission step of cytokinesis. Until recently, it was thought that abscission-regulating proteins, such as ESCRT-III complex subunits, accumulate at the MB by directly or indirectly binding to the MB resident protein, CEP55. However, recent studies have shown that depletion of CEP55 does not fully block ESCRT-III targeting the MB. Here, we show that MBs contain mRNAs and that these MB-associated mRNAs can be locally translated, resulting in the accumulation of abscission-regulating proteins. We demonstrate that localized MB-associated translation of CHMP4B is required for its targeting to the abscission site and that 3' UTR-dependent CHMP4B mRNA targeting to the MB is required for successful completion of cytokinesis. Finally, we identify regulatory cis-elements within RNAs that are necessary and sufficient for mRNA trafficking to the MB. We propose a novel method of regulating cytokinesis and abscission by MB-associated targeting and localized translation of selective mRNAs.
© 2023 Farmer et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures








Similar articles
-
The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis.Nat Commun. 2020 Apr 22;11(1):1941. doi: 10.1038/s41467-020-15205-z. Nat Commun. 2020. PMID: 32321914 Free PMC article.
-
UMAD1 contributes to ESCRT-III dynamic subunit turnover during cytokinetic abscission.J Cell Sci. 2023 Aug 1;136(15):jcs261097. doi: 10.1242/jcs.261097. Epub 2023 Aug 10. J Cell Sci. 2023. PMID: 37439191 Free PMC article.
-
Integrin signaling via FAK-Src controls cytokinetic abscission by decelerating PLK1 degradation and subsequent recruitment of CEP55 at the midbody.Oncotarget. 2016 May 24;7(21):30820-30. doi: 10.18632/oncotarget.9003. Oncotarget. 2016. PMID: 27127172 Free PMC article.
-
Knowing when to cut and run: mechanisms that control cytokinetic abscission.Trends Cell Biol. 2013 Sep;23(9):433-41. doi: 10.1016/j.tcb.2013.04.006. Epub 2013 May 22. Trends Cell Biol. 2013. PMID: 23706391 Review.
-
Resurrecting remnants: the lives of post-mitotic midbodies.Trends Cell Biol. 2013 Mar;23(3):118-28. doi: 10.1016/j.tcb.2012.10.012. Epub 2012 Dec 11. Trends Cell Biol. 2013. PMID: 23245592 Free PMC article. Review.
Cited by
-
The midbody and midbody remnant: from cellular debris to signaling organelle with diagnostic and therapeutic potential.Mol Biol Cell. 2025 Jul 1;36(7):re4. doi: 10.1091/mbc.E25-03-0120. Epub 2025 May 28. Mol Biol Cell. 2025. PMID: 40434898 Free PMC article. Review.
-
An oocyte meiotic midbody cap is required for developmental competence in mice.Nat Commun. 2023 Nov 16;14(1):7419. doi: 10.1038/s41467-023-43288-x. Nat Commun. 2023. PMID: 37973997 Free PMC article.
-
NudCL2 is required for cytokinesis by stabilizing RCC2 with Hsp90 at the midbody.Protein Cell. 2024 Oct 1;15(10):766-782. doi: 10.1093/procel/pwae025. Protein Cell. 2024. PMID: 38801297 Free PMC article.
-
Analyzing RNA Localization Using the RNA Proximity Labeling Method OINC-seq.Bio Protoc. 2025 Aug 5;15(15):e5403. doi: 10.21769/BioProtoc.5403. eCollection 2025 Aug 5. Bio Protoc. 2025. PMID: 40799472 Free PMC article.
-
miR-31-mediated local translation at the mitotic spindle is important for early development.Development. 2024 Sep 1;151(17):dev202619. doi: 10.1242/dev.202619. Epub 2024 Sep 5. Development. 2024. PMID: 39250531 Free PMC article.
References
-
- Addi, C., Presle A., Frémont S., Cuvelier F., Rocancourt M., Milin F., Schmutz S., Chamot-Rooke J., Douché T., Duchateau M., et al. . 2020. The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis. Nat. Commun. 11:1941. 10.1038/s41467-020-15205-z - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

