Hypoxia increases persulfide and polysulfide formation by AMP kinase dependent cystathionine gamma lyase phosphorylation
- PMID: 37922764
- PMCID: PMC10641705
- DOI: 10.1016/j.redox.2023.102949
Hypoxia increases persulfide and polysulfide formation by AMP kinase dependent cystathionine gamma lyase phosphorylation
Abstract
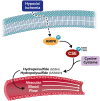
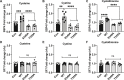
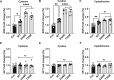
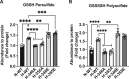
Hydropersulfide and hydropolysulfide metabolites are increasingly important reactive sulfur species (RSS) regulating numerous cellular redox dependent functions. Intracellular production of these species is known to occur through RSS interactions or through translational mechanisms involving cysteinyl t-RNA synthetases. However, regulation of these species under cell stress conditions, such as hypoxia, that are known to modulate RSS remain poorly understood. Here we define an important mechanism of increased persulfide and polysulfide production involving cystathionine gamma lyase (CSE) phosphorylation at serine 346 and threonine 355 in a substrate specific manner, under acute hypoxic conditions. Hypoxic phosphorylation of CSE occurs in an AMP kinase dependent manner increasing enzyme activity involving unique inter- and intramolecular interactions within the tetramer. Importantly, both cellular hypoxia and tissue ischemia result in AMP Kinase dependent CSE phosphorylation that regulates blood flow in ischemic tissues. Our findings reveal hypoxia molecular signaling pathways regulating CSE dependent persulfide and polysulfide production impacting tissue and cellular response to stress.
Keywords: AMP kinase; Cystathionine gamma lyase; Ischemia; Molecular modeling; Persulfide; Phosphorylation; Polysulfide.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest C.G.K., G.K·K., S.A., and X.S. have a provisional patent on CSE phosphorylation mutants and uses.
Figures




References
-
- Fukuto J.M., Ignarro L.J., Nagy P., Wink D.A., Kevil C.G., Feelisch M., Cortese-Krott M.M., Bianco C.L., Kumagai Y., Hobbs A.J., et al. Biological hydropersulfides and related polysulfides - a new concept and perspective in redox biology. FEBS Lett. 2018;592:2140–2152. doi: 10.1002/1873-3468.13090. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

