Envisioning a new era: Complete genetic information from routine, telomere-to-telomere genomes
- PMID: 37922882
- PMCID: PMC10645551
- DOI: 10.1016/j.ajhg.2023.09.011
Envisioning a new era: Complete genetic information from routine, telomere-to-telomere genomes
Abstract
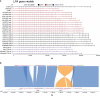
Advances in long-read sequencing and assembly now mean that individual labs can generate phased genomes that are more accurate and more contiguous than the original human reference genome. With declining costs and increasing democratization of technology, we suggest that complete genome assemblies, where both parental haplotypes are phased telomere to telomere, will become standard in human genetics. Soon, even in clinical settings where rigorous sample-handling standards must be met, affected individuals could have reference-grade genomes fully sequenced and assembled in just a few hours given advances in technology, computational processing, and annotation. Complete genetic variant discovery will transform how we map, catalog, and associate variation with human disease and fundamentally change our understanding of the genetic diversity of all humans.
Copyright © 2023 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.E.E. is a scientific advisory board (SAB) member of Variant Bio, Inc.
Figures

Comment in
-
To boldly go: Unpacking the NHGRI's bold predictions for human genomics by 2030.Am J Hum Genet. 2023 Nov 2;110(11):1829-1831. doi: 10.1016/j.ajhg.2023.09.010. Am J Hum Genet. 2023. PMID: 37922881 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

