Sequencing-based functional assays for classification of BRCA2 variants in mouse ESCs
- PMID: 37922907
- PMCID: PMC10694496
- DOI: 10.1016/j.crmeth.2023.100628
Sequencing-based functional assays for classification of BRCA2 variants in mouse ESCs
Abstract
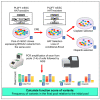
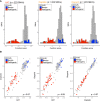
Sequencing of genes, such as BRCA1 and BRCA2, is recommended for individuals with a personal or family history of early onset and/or bilateral breast and/or ovarian cancer or a history of male breast cancer. Such sequencing efforts have resulted in the identification of more than 17,000 BRCA2 variants. The functional significance of most variants remains unknown; consequently, they are called variants of uncertain clinical significance (VUSs). We have previously developed mouse embryonic stem cell (mESC)-based assays for functional classification of BRCA2 variants. We now developed a next-generation sequencing (NGS)-based approach for functional evaluation of BRCA2 variants using pools of mESCs expressing 10-25 BRCA2 variants from a given exon. We use this approach for functional evaluation of 223 variants listed in ClinVar. Our functional classification of BRCA2 variants is concordant with the classification reported in ClinVar or those reported by other orthogonal assays.
Keywords: BAC; BRCA2; CP: Cancer biology; CP: Genetics; DNA repair; VUS; bacterial artificial chromosome; breast cancer; cell viability; functional assay; mouse ES Cells; recombineering; variants of uncertain significance.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
Saturation genome editing of 11 codons and exon 13 of BRCA2 coupled with chemotherapeutic drug response accurately determines pathogenicity of variants.PLoS Genet. 2023 Sep 15;19(9):e1010940. doi: 10.1371/journal.pgen.1010940. eCollection 2023 Sep. PLoS Genet. 2023. PMID: 37713444 Free PMC article.
-
DNA repair-related functional assays for the classification of BRCA1 and BRCA2 variants: a critical review and needs assessment.J Med Genet. 2017 Nov;54(11):721-731. doi: 10.1136/jmedgenet-2017-104707. Epub 2017 Sep 2. J Med Genet. 2017. PMID: 28866612 Review.
-
Epidemiological and ES cell-based functional evaluation of BRCA2 variants identified in families with breast cancer.Hum Mutat. 2021 Feb;42(2):200-212. doi: 10.1002/humu.24154. Epub 2020 Dec 31. Hum Mutat. 2021. PMID: 33314489 Free PMC article.
-
Functional evaluation of five BRCA2 unclassified variants identified in a Sri Lankan cohort with inherited cancer syndromes using a mouse embryonic stem cell-based assay.Breast Cancer Res. 2020 May 11;22(1):43. doi: 10.1186/s13058-020-01272-z. Breast Cancer Res. 2020. PMID: 32393398 Free PMC article.
-
Functional assays for analysis of variants of uncertain significance in BRCA2.Hum Mutat. 2014 Feb;35(2):151-64. doi: 10.1002/humu.22478. Epub 2013 Dec 3. Hum Mutat. 2014. PMID: 24323938 Free PMC article. Review.
Cited by
-
Saturation genome editing-based clinical classification of BRCA2 variants.Nature. 2025 Feb;638(8050):538-545. doi: 10.1038/s41586-024-08349-1. Epub 2025 Jan 8. Nature. 2025. PMID: 39779848
References
-
- LaDuca H., Polley E.C., Yussuf A., Hoang L., Gutierrez S., Hart S.N., Yadav S., Hu C., Na J., Goldgar D.E., et al. A clinical guide to hereditary cancer panel testing: evaluation of gene-specific cancer associations and sensitivity of genetic testing criteria in a cohort of 165,000 high-risk patients. Genet. Med. 2020;22:407–415. doi: 10.1038/s41436-019-0633-8. - DOI - PMC - PubMed
-
- Paluch-Shimon S., Cardoso F., Sessa C., Balmana J., Cardoso M.J., Gilbert F., Senkus E., ESMO Guidelines Committee Prevention and screening in BRCA mutation carriers and other breast/ovarian hereditary cancer syndromes: ESMO Clinical Practice Guidelines for cancer prevention and screening. Ann. Oncol. 2016;27:v103–v110. doi: 10.1093/annonc/mdw327. - DOI - PubMed
-
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. - DOI - PMC - PubMed
-
- Brnich S.E., Abou Tayoun A.N., Couch F.J., Cutting G.R., Greenblatt M.S., Heinen C.D., Kanavy D.M., Luo X., McNulty S.M., Starita L.M., et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019;12:3. doi: 10.1186/s13073-019-0690-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

