Domain-Selective BET Ligands Yield Next-Generation Synthetic Genome Readers/Regulators with Nonidentical Cellular Functions
- PMID: 37923569
- PMCID: PMC11957383
- DOI: 10.1021/jacs.3c06297
Domain-Selective BET Ligands Yield Next-Generation Synthetic Genome Readers/Regulators with Nonidentical Cellular Functions
Abstract
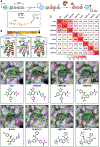
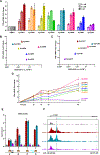
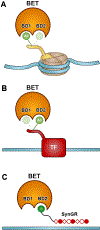
SynTEF1, a prototype synthetic genome reader/regulator (SynGR), was designed to target GAA triplet repeats and restore the expression of frataxin (FXN) in Friedreich's ataxia patients. It achieves this complex task by recruiting BRD4, via a pan-BET ligand (JQ1), to the GAA repeats by using a sequence-selective DNA-binding polyamide. When bound to specific genomic loci in this way, JQ1 functions as a chemical prosthetic for acetyl-lysine residues that are natural targets of the two tandem bromodomains (BD1 and BD2) in bromo- and extra-terminal domain (BET) proteins. As next-generation BET ligands were disclosed, we tested a select set with improved physicochemical, pharmacological, and bromodomain-selective properties as substitutes for JQ1 in the SynGR design. Here, we report two unexpected findings: (1) SynGRs bearing pan-BET or BD2-selective ligands license transcription at the FXN locus, whereas those bearing BD1-selective ligands do not, and (2) rather than being neutral or inhibitory, an untethered BD1-selective ligand (GSK778) substantively enhances the activity of all active SynGRs. The failure of BD1-selective SynGRs to recruit BRD4/BET proteins suggests that rather than functioning as "epigenetic/chromatin mimics," active SynGRs mimic the functions of natural transcription factors in engaging BET proteins through BD2 binding. Moreover, the enhanced activity of SynGRs upon cotreatment with the BD1-selective ligand suggests that natural transcription factors compete for a limited pool of nonchromatin-bound BET proteins, and blocking BD1 directs pan-BET ligands to more effectively engage BD2. Taken together, SynGRs as chemical probes provide unique insights into the molecular recognition principles utilized by natural factors to precisely regulate gene expression, and they guide the design of more sophisticated synthetic gene regulators with greater therapeutic potential.
Figures


References
-
- Dervan PB, A Personal Perspective on Chemical Biology: Before the Beginning. Israel Journal of Chemistry 2019, 59 (1–2), 71–83.
-
- Ptashne M; Gann A, Genes & signals. Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY:: 2002; Vol. 402.
-
- Henley MJ; Koehler AN, Advances in targeting ‘undruggable’transcription factors with small molecules. Nature Reviews Drug Discovery 2021, 20 (9), 669–688. - PubMed
-
- White S; Szewczyk JW; Turner JM; Baird EE; Dervan PB, Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature 1998, 391 (6666), 468–71. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

