Structure of a membrane-bound menaquinol:organohalide oxidoreductase
- PMID: 37923808
- PMCID: PMC10624902
- DOI: 10.1038/s41467-023-42927-7
Structure of a membrane-bound menaquinol:organohalide oxidoreductase
Abstract
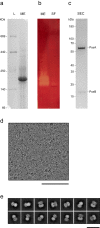
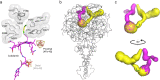
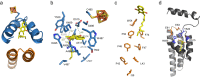
Organohalide-respiring bacteria are key organisms for the bioremediation of soils and aquifers contaminated with halogenated organic compounds. The major players in this process are respiratory reductive dehalogenases, corrinoid enzymes that use organohalides as substrates and contribute to energy conservation. Here, we present the structure of a menaquinol:organohalide oxidoreductase obtained by cryo-EM. The membrane-bound protein was isolated from Desulfitobacterium hafniense strain TCE1 as a PceA2B2 complex catalysing the dechlorination of tetrachloroethene. Two catalytic PceA subunits are anchored to the membrane by two small integral membrane PceB subunits. The structure reveals two menaquinone molecules bound at the interface of the two different subunits, which are the starting point of a chain of redox cofactors for electron transfer to the active site. In this work, the structure elucidates how energy is conserved during organohalide respiration in menaquinone-dependent organohalide-respiring bacteria.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 1998;22:383–398. doi: 10.1111/j.1574-6976.1998.tb00377.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

