Six-hour time-restricted feeding inhibits lung cancer progression and reshapes circadian metabolism
- PMID: 37924048
- PMCID: PMC10625271
- DOI: 10.1186/s12916-023-03131-y
Six-hour time-restricted feeding inhibits lung cancer progression and reshapes circadian metabolism
Abstract
Background: Accumulating evidence has suggested an oncogenic effect of diurnal disruption on cancer progression. To test whether targeting circadian rhythm by dietary strategy suppressed lung cancer progression, we adopted 6-h time-restricted feeding (TRF) paradigm to elucidate whether and how TRF impacts lung cancer progression.
Methods: This study used multiple lung cancer cell lines, two xenograft mouse models, and a chemical-treated mouse lung cancer model. Stable TIM-knockdown and TIM-overexpressing A549 cells were constructed. Cancer behaviors in vitro were determined by colony formation, EdU proliferation, wound healing, transwell migration, flow cytometer, and CCK8 assays. Immunofluorescence, pathology examinations, and targeted metabolomics were also used in tumor cells and tissues. mCherry-GFP-LC3 plasmid was used to detect autophagic flux.
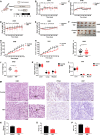
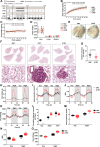
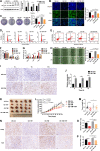
Results: We found for the first time that compared to normal ad libitum feeding, 6-h TRF inhibited lung cancer progression and reprogrammed the rhythms of metabolites or genes involved in glycolysis and the circadian rhythm in tumors. After TRF intervention, only timeless (TIM) gene among five lung cancer-associated clock genes was found to consistently align rhythm of tumor cells to that of tumor tissues. Further, we demonstrated that the anti-tumor effect upon TRF was partially mediated by the rhythmic downregulation of the TIM and the subsequent activation of autophagy. Combining TRF with TIM inhibition further enhanced the anti-tumor effect, comparable to treatment efficacy of chemotherapy in xenograft model.
Conclusions: Six-hour TRF inhibits lung cancer progression and reshapes circadian metabolism, which is partially mediated by the rhythmic downregulation of the TIM and the subsequent upregulation of autophagy.
Keywords: Autophagy; Circadian rhythm; Lung cancer; Progression; TIM; Time-restricted feeding.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflict of interests.
Figures





References
-
- Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, Bonaventure A, Valkov M, Johnson CJ, Esteve J, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. - DOI - PMC - PubMed
-
- Pariollaud M, Lamia KA. Cancer in the fourth dimension: what is the impact of circadian disruption? Cancer Discov. 2020;10(10):1455–1464. doi: 10.1158/2159-8290.CD-20-0413. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

