A combined adjuvant approach primes robust germinal center responses and humoral immunity in non-human primates
- PMID: 37925510
- PMCID: PMC10625619
- DOI: 10.1038/s41467-023-42923-x
A combined adjuvant approach primes robust germinal center responses and humoral immunity in non-human primates
Abstract
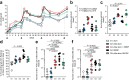
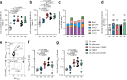
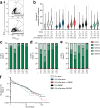
Adjuvants and antigen delivery kinetics can profoundly influence B cell responses and should be critically considered in rational vaccine design, particularly for difficult neutralizing antibody targets such as human immunodeficiency virus (HIV). Antigen kinetics can change depending on the delivery method. To promote extended immunogen bioavailability and to present antigen in a multivalent form, native-HIV Env trimers are modified with short phosphoserine peptide linkers that promote tight binding to aluminum hydroxide (pSer:alum). Here we explore the use of a combined adjuvant approach that incorporates pSer:alum-mediated antigen delivery with potent adjuvants (SMNP, 3M-052) in an extensive head-to-head comparison study with conventional alum to assess germinal center (GC) and humoral immune responses. Priming with pSer:alum plus SMNP induces additive effects that enhance the magnitude and persistence of GCs, which correlate with better GC-TFH cell help. Autologous HIV-neutralizing antibody titers are improved in SMNP-immunized animals after two immunizations. Over 9 months after priming immunization of pSer:alum with either SMNP or 3M-052, robust Env-specific bone marrow plasma cells (BM BPC) are observed. Furthermore, pSer-modification of Env trimer reduce targeting towards immunodominant non-neutralizing epitopes. The study shows that a combined adjuvant approach can augment humoral immunity by modulating immunodominance and shows promise for clinical translation.
© 2023. The Author(s).
Conflict of interest statement
W.R.S is named as an inventor on a patent for the MD39 immunogen (US No. 11,203,617). D.J.I. and W.R.S. are named as inventors on a patent for pSer technology (US No. 11,224,648 B2). D.J.I and S.C. are inventors on a patent for the SMNP adjuvant (US No. 11,547,672 B2). K.A.R. and D.J.I. are inventors on patent applications for the synergistic combination of alum and SMNP adjuvants (PCT/US2022/074302 and US No. 17/816,045). The remaining authors declare no competing interests.
Figures



Similar articles
-
Vaccines combining slow release and follicle targeting of antigens increase germinal center B cell diversity and clonal expansion.Sci Transl Med. 2025 Jun 18;17(803):eadw7499. doi: 10.1126/scitranslmed.adw7499. Epub 2025 Jun 18. Sci Transl Med. 2025. PMID: 40531969 Free PMC article.
-
Engineered immunogen binding to alum adjuvant enhances humoral immunity.Nat Med. 2020 Mar;26(3):430-440. doi: 10.1038/s41591-020-0753-3. Epub 2020 Feb 17. Nat Med. 2020. PMID: 32066977 Free PMC article.
-
Early T Follicular Helper Cell Responses and Germinal Center Reactions Are Associated with Viremia Control in Immunized Rhesus Macaques.J Virol. 2019 Feb 5;93(4):e01687-18. doi: 10.1128/JVI.01687-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30463978 Free PMC article.
-
Vaccines combining slow delivery and follicle targeting of antigens increase germinal center B cell clonal diversity and clonal expansion.bioRxiv [Preprint]. 2024 Aug 19:2024.08.19.608655. doi: 10.1101/2024.08.19.608655. bioRxiv. 2024. Update in: Sci Transl Med. 2025 Jun 18;17(803):eadw7499. doi: 10.1126/scitranslmed.adw7499. PMID: 39229011 Free PMC article. Updated. Preprint.
-
Tfh cells and HIV bnAbs, an immunodominance model of the HIV neutralizing antibody generation problem.Immunol Rev. 2017 Jan;275(1):49-61. doi: 10.1111/imr.12512. Immunol Rev. 2017. PMID: 28133798 Review.
Cited by
-
Modulation of antigen delivery and lymph node activation in non-human primates by saponin adjuvant SMNP.bioRxiv [Preprint]. 2024 Aug 28:2024.08.28.608716. doi: 10.1101/2024.08.28.608716. bioRxiv. 2024. Update in: PNAS Nexus. 2024 Nov 25;3(12):pgae529. doi: 10.1093/pnasnexus/pgae529. PMID: 39253464 Free PMC article. Updated. Preprint.
-
Cancer Vaccines and Beyond: The Transformative Role of Nanotechnology in Immunotherapy.Pharmaceutics. 2025 Feb 7;17(2):216. doi: 10.3390/pharmaceutics17020216. Pharmaceutics. 2025. PMID: 40006583 Free PMC article. Review.
-
Vaccination with an mRNA-encoded membrane-bound HIV envelope trimer induces neutralizing antibodies in animal models.Sci Transl Med. 2025 Jul 30;17(809):eadw0721. doi: 10.1126/scitranslmed.adw0721. Epub 2025 Jul 30. Sci Transl Med. 2025. PMID: 40737430 Free PMC article.
-
Vaccination with mRNA-encoded membrane-bound HIV Envelope trimer induces neutralizing antibodies in animal models.bioRxiv [Preprint]. 2025 Jan 25:2025.01.24.634423. doi: 10.1101/2025.01.24.634423. bioRxiv. 2025. Update in: Sci Transl Med. 2025 Jul 30;17(809):eadw0721. doi: 10.1126/scitranslmed.adw0721. PMID: 39896562 Free PMC article. Updated. Preprint.
-
Modulation of antigen delivery and lymph node activation in nonhuman primates by saponin adjuvant saponin/monophosphoryl lipid A nanoparticle.PNAS Nexus. 2024 Nov 25;3(12):pgae529. doi: 10.1093/pnasnexus/pgae529. eCollection 2024 Dec. PNAS Nexus. 2024. PMID: 39677368 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

