Alternative lengthening of telomeres (ALT) cells viability is dependent on C-rich telomeric RNAs
- PMID: 37925537
- PMCID: PMC10625592
- DOI: 10.1038/s41467-023-42831-0
Alternative lengthening of telomeres (ALT) cells viability is dependent on C-rich telomeric RNAs
Abstract
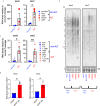
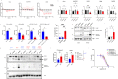
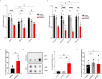
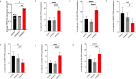
Alternative lengthening of telomeres (ALT) is a telomere maintenance mechanism activated in ~10-15% of cancers, characterized by telomeric damage. Telomeric damage-induced long non-coding RNAs (dilncRNAs) are transcribed at dysfunctional telomeres and contribute to telomeric DNA damage response (DDR) activation and repair. Here we observed that telomeric dilncRNAs are preferentially elevated in ALT cells. Inhibition of C-rich (teloC) dilncRNAs with antisense oligonucleotides leads to DNA replication stress responses, increased genomic instability, and apoptosis induction selectively in ALT cells. Cell death is dependent on DNA replication and is increased by DNA replication stress. Mechanistically, teloC dilncRNA inhibition reduces RAD51 and 53BP1 recruitment to telomeres, boosts the engagement of BIR machinery, and increases C-circles and telomeric sister chromatid exchanges, without increasing telomeric non-S phase synthesis. These results indicate that teloC dilncRNA is necessary for a coordinated recruitment of DDR factors to ALT telomeres and it is essential for ALT cancer cells survival.
© 2023. The Author(s).
Conflict of interest statement
F.R. and F.d.A.d.F. are inventors on the patent applications PCT/EP2013/059753 and PCT/EP2016/068162. C.J.W. and J.A. are inventors on the patent application PCT/ EP2016/068162. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

