Significant variation of filamentation phenotypes in clinical Candida albicans strains
- PMID: 37928181
- PMCID: PMC10623444
- DOI: 10.3389/fcimb.2023.1207083
Significant variation of filamentation phenotypes in clinical Candida albicans strains
Abstract
Introduction: Candida albicans is an opportunistic human pathogen that typically resides as part of the microbiome in the gastrointestinal and genitourinary tracts of a large portion of the human population. This fungus lacks a true sexual cycle and evolves in a largely clonal pattern. The ability to cause disease is consistent across the species as strains causing systemic infections appear across the known C. albicans intra-species clades.
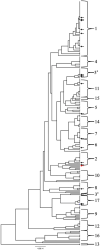
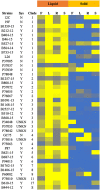
Methods: In this work, strains collected from patients with systemic C. albicans infections isolated at the Nebraska Medicine clinical laboratory were typed by MLST analysis. Since the ability to form filaments has been linked to pathogenesis in C. albicans, these clinical strains, as well as a previously genotyped set of clinical strains, were tested for their ability to filament across a variety of inducing conditions.
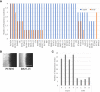
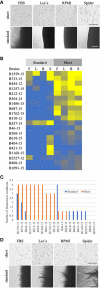
Results: Genotyping of the clinical strains demonstrated that the strains isolated at one of the major medical centers in our region were as diverse as strains collected across the United States. We demonstrated that clinical strains exhibit a variety of filamentation patterns across differing inducing conditions. The only consistent pattern observed in the entire set of clinical strains tested was an almost universal inability to filament in standard solid inducing conditions used throughout the C. albicans field. A different solid filamentation assay that produces more robust filamentation profiles from clinical strains is proposed in this study, although not all strains expected to filament in vivo were filamentous in this assay.
Discussion: Our data supports growing evidence that broad phenotypic diversity exists between the C. albicans type strain and clinical strains, suggesting that the type strain poorly represents filamentation patterns observed in most clinical isolates. These data further highlight the need to use diverse clinical strains in pathogenesis assays.
Keywords: Candida albicans; clinical strains; filamentation; hyphal morphogenesis; phylogeny.
Copyright © 2023 Brandquist, Lampman, Smith, Basilio, Almansob, Iwen and Blankenship.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Cavalieri D., Di Paola M., Rizzetto L., Tocci N., De Filippo C., Lionetti P., et al. . (2017). Genomic and phenotypic variation in morphogenetic networks of two Candida albicans isolates subtends their different pathogenic potential. Front. Immunol. 8, 1997. doi: 10.3389/fimmu.2017.01997 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

