Mesoscope: A Web-based Tool for Mesoscale Data Integration and Curation
- PMID: 37928321
- PMCID: PMC10624244
- DOI: 10.2312/molva.20201098
Mesoscope: A Web-based Tool for Mesoscale Data Integration and Curation
Abstract
Interest is growing for 3D models of the biological mesoscale, the intermediate scale between the nanometer scale of molecular structure and micrometer scale of cellular biology. However, it is currently difficult to gather, curate and integrate all the data required to define such models. To address this challenge we developed Mesoscope (mesoscope.scripps.edu/beta), a web-based data integration and curation tool. Mesoscope allows users to begin with a listing of molecules (such as data from proteomics), and to use resources at UniProt and the PDB to identify, prepare and validate appropriate structures and representations for each molecule, ultimately producing a portable output file used by CellPACK and other modeling tools for generation of 3D models of the biological mesoscale. The availability of this tool has proven essential in several exploratory applications, given the high complexity of mesoscale models and the heterogeneity of the available data sources.
Keywords: Applied computing → Bioinformatics; Computing methodologies → Graphics systems and interfaces.
Figures
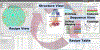


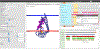






References
-
- DANA JM, GUTMANAS A, TYAGI N, QI G, O’DONOVAN C, MARTIN M, VELANKAR S: SIFTS: updated Structure Integration with Function, Taxonomy and Sequences resource allows 40-fold increase in coverage of structure-based annotations for proteins. Nucleic Acids Research 47, D1 (11 2018), D482–D489. URL: 10.1093/nar/gky1114, arXiv:https://academic.oup.com/nar/article-pdf/47/D1/D482/27437256/gky11..., doi:10.1093/nar/gky1114.4 - DOI - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
