Prevalence of diagnostically-discrepant Clostridioides difficile clinical specimens: insights from longitudinal surveillance
- PMID: 37928470
- PMCID: PMC10622765
- DOI: 10.3389/fmed.2023.1238159
Prevalence of diagnostically-discrepant Clostridioides difficile clinical specimens: insights from longitudinal surveillance
Abstract
Background: Clostridioides difficile Infection (CDI) is a healthcare-associated diarrheal disease prevalent worldwide. A common diagnostic algorithm relies on a two-step protocol that employs stool enzyme immunoassays (EIAs) to detect the pathogen, and its toxins, respectively. Active CDI is deemed less likely when the Toxin EIA result is negative, even if the pathogen-specific EIA is positive for C. difficile. We recently reported, however, that low-toxin-producing C. difficile strains recovered from Toxin-negative ('discrepant') clinical stool specimens can be fully pathogenic, and cause lethality in a rodent CDI model. To document frequency of discrepant CDI specimens, and evaluate C. difficile strain diversity, we performed longitudinal surveillance at a Southern Arizona tertiary-care hospital.
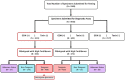
Methods: Diarrheic stool specimens from patients with clinical suspicion of CDI were obtained over an eight-year period (2015-2022) from all inpatient and outpatient Units of a > 600-bed Medical Center in Southern Arizona. Clinical laboratory EIA testing identified C. difficile-containing specimens, and classified them as Toxin-positive or Toxin-negative. C. difficile isolates recovered from the stool specimens were DNA fingerprinted using an international phylogenetic lineage assignment system ("ribotyping"). For select isolates, toxin abundance in stationary phase supernatants of pure cultures was quantified via EIA.
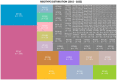
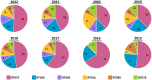
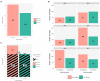
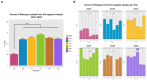
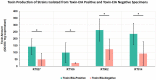
Results: Of 8,910 diarrheic specimens that underwent diagnostic testing, 1733 (19.4%) harbored C. difficile. Our major findings were that: (1) C. difficile prevalence and phylogenetic diversity was stable over the 8-year period; (2) toxigenic C. difficile was recovered from 69% of clinically Tox-neg ('discrepant') specimens; (3) the six most prevalent USA ribotypes were recovered in significant proportions (>60%) from Tox-neg specimens; and (4) toxin-producing C. difficile recovered from discrepant specimens produced less toxin than strains of the same ribotype isolated from non-discrepant specimens.
Conclusion: Our study highlights the dominance of Toxin EIA-negative CDI specimens in a clinical setting and the high frequency of known virulent ribotypes in these specimens. Therefore, a careful reevaluation of the clinical relevance of diagnostically-discrepant specimens particularly in the context of missed CDI diagnoses and C. difficile persistence, is warranted.
Keywords: Clostridioides difficile; discrepant; longitudinal; prevalence; ribotyping; surveillance.
Copyright © 2023 Anwar, Clark, Lindsey, Claus-Walker, Mansoor, Nguyen, Billy, Lainhart, Shehab, Viswanathan and Vedantam.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Low-toxin Clostridioides difficile RT027 strains exhibit robust virulence.Emerg Microbes Infect. 2022 Dec;11(1):1982-1993. doi: 10.1080/22221751.2022.2105260. Emerg Microbes Infect. 2022. PMID: 35880487 Free PMC article.
-
Performance of laboratory tests for detection for Clostridioides difficile: A multicenter prospective study in Japan.Anaerobe. 2019 Dec;60:102107. doi: 10.1016/j.anaerobe.2019.102107. Epub 2019 Oct 21. Anaerobe. 2019. PMID: 31647977
-
Changes in the Association Between Diagnostic Testing Method, Polymerase Chain Reaction Ribotype, and Clinical Outcomes From Clostridioides difficile Infection: One Institution's Experience.Clin Infect Dis. 2021 Nov 2;73(9):e2883-e2889. doi: 10.1093/cid/ciaa1395. Clin Infect Dis. 2021. PMID: 32930705 Free PMC article.
-
Laboratory Diagnostic Methods for Clostridioides difficile Infection: the First Systematic Review and Meta-analysis in Korea.Ann Lab Med. 2021 Mar 1;41(2):171-180. doi: 10.3343/alm.2021.41.2.171. Ann Lab Med. 2021. PMID: 33063678 Free PMC article.
-
Insights into the Evolving Epidemiology of Clostridioides difficile Infection and Treatment: A Global Perspective.Antibiotics (Basel). 2023 Jul 1;12(7):1141. doi: 10.3390/antibiotics12071141. Antibiotics (Basel). 2023. PMID: 37508237 Free PMC article. Review.
Cited by
-
Clostridioides difficile superoxide reductase mitigates oxygen sensitivity.J Bacteriol. 2024 Jul 25;206(7):e0017524. doi: 10.1128/jb.00175-24. Epub 2024 Jul 2. J Bacteriol. 2024. PMID: 38953644 Free PMC article.
References
-
- CDC . Antibiotic resistance threats in the United States. Atlanta, GA: U.S. Department of Health and Human Services, CDC. (2019).
Grants and funding
LinkOut - more resources
Full Text Sources

