Plasma proteomic profiling of bacterial cold water disease-resistant and -susceptible rainbow trout lines and biomarker discovery
- PMID: 37928534
- PMCID: PMC10623068
- DOI: 10.3389/fimmu.2023.1265386
Plasma proteomic profiling of bacterial cold water disease-resistant and -susceptible rainbow trout lines and biomarker discovery
Abstract
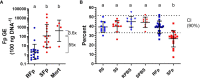
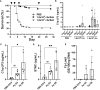
Genetic variation for disease resistance is present in salmonid fish; however, the molecular basis is poorly understood, and biomarkers of disease susceptibility/resistance are unavailable. Previously, we selected a line of rainbow trout for high survival following standardized challenge with Flavobacterium psychrophilum (Fp), the causative agent of bacterial cold water disease. The resistant line (ARS-Fp-R) exhibits over 60 percentage points higher survival compared to a reference susceptible line (ARS-Fp-S). To gain insight into the differential host response between genetic lines, we compared the plasma proteomes from day 6 after intramuscular challenge. Pooled plasma from unhandled, PBS-injected, and Fp-injected groups were simultaneously analyzed using a TMT 6-plex label, and the relative abundance of 513 proteins was determined. Data are available via ProteomeXchange, with identifier PXD041308, and the relative protein abundance values were compared to mRNA measured from a prior, whole-body RNA-seq dataset. Our results identified a subset of differentially abundant intracellular proteins was identified, including troponin and myosin, which were not transcriptionally regulated, suggesting that these proteins were released into plasma following pathogen-induced tissue damage. A separate subset of high-abundance, secreted proteins were transcriptionally regulated in infected fish. The highest differentially expressed protein was a C1q family member (designated complement C1q-like protein 3; C1q-LP3) that was upregulated over 20-fold in the infected susceptible line while only modestly upregulated, 1.8-fold, in the infected resistant line. Validation of biomarkers was performed using immunoassays and C1q-LP3, skeletal muscle troponin C, cathelcidin 2, haptoglobin, leptin, and growth and differentiation factor 15 exhibited elevated concentration in susceptible line plasma. Complement factor H-like 1 exhibited higher abundance in the resistant line compared to the susceptible line in both control and challenged fish and thus was a baseline differentiator between lines. C1q-LP3 and STNC were elevated in Atlantic salmon plasma following experimental challenge with Fp. In summary, these findings further the understanding of the differential host response to Fp and identifies salmonid biomarkers that may have use for genetic line evaluation and on-farm health monitoring.
Keywords: Flavobacterium psychrophilum; bacterial cold water disease; biomarker; complement C1q-like protein 3; complement factor H-like 1; disease resistance.
Copyright © 2023 Wiens, Marancik, Chadwick, Osbourn, Reid and Leeds.
Conflict of interest statement
CC is an owner and employed by Life Diagnostics, and co-owner of Veterinary Biomarkers. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Embody GC, Hyford CD. The advantage of rearing brook trout fingerlings from selected breeders. Trans Amer Fish Soc (1925) 55):135–8. doi: 10.1577/1548-8659(1925)55[135:TAORB - DOI
-
- Wolf LE. Development of disease-resistant strains of fish. Trans Am Fish Soc (1954) 83(1):342–9. doi: 10.1577/1548-8659(1953)83[342:DODSO - DOI
-
- Fjalestad KT, Gjedrem T, Gjerde B. Genetic improvement of disease resistance in fish: an overview. Aquaculture (1993) 111:65–74. doi: 10.1016/B978-0-444-81527-9.50011-7 - DOI
-
- Gjedrem T. Selection and breeding programs in aquaculture. Dordrecht, Netherlands: Springer; (2005). Available at: https://link.springer.com/book/10.1007/1-4020-3342-7.
-
- Gjedrem T. Disease resistant fish and shellfish are within reach: A review. J Mar Sci Eng (2015) 3:146–53. doi: 10.3390/jmse3010146 - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

