SARS-CoV-2 NSP12 associates with TRiC and the P323L substitution acts as a host adaption
- PMID: 37929963
- PMCID: PMC10688337
- DOI: 10.1128/jvi.00424-23
SARS-CoV-2 NSP12 associates with TRiC and the P323L substitution acts as a host adaption
Abstract
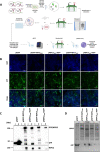
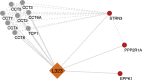
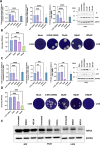
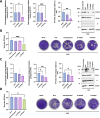
SARS-CoV-2 has caused a worldwide health and economic crisis. During the course of the pandemic, genetic changes occurred in the virus, which have resulted in new properties of the virus-particularly around gains in transmission and the ability to partially evade either natural or vaccine-acquired immunity. Some of these viruses have been labeled Variants of Concern (VoCs). At the root of all VoCs are two mutations, one in the viral spike protein that has been very well characterized and the other in the virus polymerase (NSP12). This is the viral protein responsible for replicating the genome. We show that NSP12 associates with host cell proteins that act as a scaffold to facilitate the function of this protein. Furthermore, we found that different variants of NSP12 interact with host cell proteins in subtle and different ways, which affect function.
Keywords: NSP12; P323L; SARS-CoV-2; TRiC; phosphatase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
SARS-CoV-2 variants with NSP12 P323L/G671S mutations display enhanced virus replication in ferret upper airways and higher transmissibility.Cell Rep. 2023 Sep 26;42(9):113077. doi: 10.1016/j.celrep.2023.113077. Epub 2023 Sep 6. Cell Rep. 2023. PMID: 37676771 Free PMC article.
-
Genomic characterization of SARS-CoV-2 isolates from patients in Turkey reveals the presence of novel mutations in spike and nsp12 proteins.J Med Virol. 2021 Oct;93(10):6016-6026. doi: 10.1002/jmv.27188. Epub 2021 Jul 16. J Med Virol. 2021. PMID: 34241906 Free PMC article.
-
Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19).2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 34033342 Free Books & Documents.
-
Characterization of SARS-CoV-2 different variants and related morbidity and mortality: a systematic review.Eur J Med Res. 2021 Jun 8;26(1):51. doi: 10.1186/s40001-021-00524-8. Eur J Med Res. 2021. PMID: 34103090 Free PMC article.
-
Mutations of SARS-CoV-2 spike protein: Implications on immune evasion and vaccine-induced immunity.Semin Immunol. 2021 Jun;55:101533. doi: 10.1016/j.smim.2021.101533. Epub 2021 Nov 20. Semin Immunol. 2021. PMID: 34836774 Free PMC article. Review.
Cited by
-
Comparative Proteomics and Interactome Analysis of the SARS-CoV-2 Nucleocapsid Protein in Human and Bat Cell Lines.Viruses. 2024 Jul 11;16(7):1117. doi: 10.3390/v16071117. Viruses. 2024. PMID: 39066279 Free PMC article.
-
The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19.bioRxiv [Preprint]. 2024 Dec 20:2024.02.27.582110. doi: 10.1101/2024.02.27.582110. bioRxiv. 2024. Update in: Microbiol Spectr. 2025 May 6;13(5):e0182924. doi: 10.1128/spectrum.01829-24. PMID: 38464327 Free PMC article. Updated. Preprint.
-
Aminoacyl-tRNA synthetase interactions in SARS-CoV-2 infection.Biochem Soc Trans. 2023 Dec 20;51(6):2127-2141. doi: 10.1042/BST20230527. Biochem Soc Trans. 2023. PMID: 38108455 Free PMC article. Review.
-
Protein folding by the CCT/TRiC chaperone complex.Curr Opin Struct Biol. 2025 Apr;91:102999. doi: 10.1016/j.sbi.2025.102999. Epub 2025 Feb 5. Curr Opin Struct Biol. 2025. PMID: 39914052 Review.
-
The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19.Microbiol Spectr. 2025 May 6;13(5):e0182924. doi: 10.1128/spectrum.01829-24. Epub 2025 Mar 25. Microbiol Spectr. 2025. PMID: 40130852 Free PMC article.
References
-
- Worobey M, Levy JI, Malpica Serrano LM, Crits-Christoph A, Pekar JE, Goldstein SA, Rasmussen AL, Kraemer MU, Newman C, Koopmans MPG, Suchard MA, Wertheim JO, Lemey P, Robertson DL, Garry RF, Holmes EC, Rambaut A, Andersen KG. 2022. The huanan market was the epicenter of SARS-CoV-2 emergence. Zenodo. doi:10.5281/zenodo.6299600 - DOI - PMC - PubMed
-
- Pekar JE, Magee A, Moshiri N, Izhikevich K, Havens JL, Gangavarapu K, Serrano LMM, Crits-Christoph A, Matteson NL, Zeller M, et al. . 2022. SARS-CoV-2 emergence very likely resulted from at least two zoonotic events. Zenodo. doi:10.5281/zenodo.6291628 - DOI
-
- Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, Mirchandani D, Scharton D, Bilello JP, Ku Z, An Z, Kalveram B, Freiberg AN, Menachery VD, Xie X, Plante KS, Weaver SC, Shi PY. 2021. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592:116–121. doi:10.1038/s41586-020-2895-3 - DOI - PMC - PubMed
-
- Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, Rangarajan ES, Pan A, Vanderheiden A, Suthar MS, Li W, Izard T, Rader C, Farzan M, Choe H. 2020. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun 11:6013. doi:10.1038/s41467-020-19808-4 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

