Tuning the potency and selectivity of ImmTAC molecules by affinity modulation
- PMID: 37930865
- PMCID: PMC10847821
- DOI: 10.1093/cei/uxad120
Tuning the potency and selectivity of ImmTAC molecules by affinity modulation
Abstract
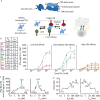
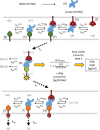
T-cell-engaging bispecifics have great clinical potential for the treatment of cancer and infectious diseases. The binding affinity and kinetics of a bispecific molecule for both target and T-cell CD3 have substantial effects on potency and specificity, but the rules governing these relationships are not fully understood. Using immune mobilizing monoclonal TCRs against cancer (ImmTAC) molecules as a model, we explored the impact of altering affinity for target and CD3 on the potency and specificity of the redirected T-cell response. This class of bispecifics binds specific target peptides presented by human leukocyte antigen on the cell surface via an affinity-enhanced T-cell receptor and can redirect T-cell activation with an anti-CD3 effector moiety. The data reveal that combining a strong affinity TCR with an intermediate affinity anti-CD3 results in optimal T-cell activation, while strong affinity of both targeting and effector domains significantly reduces maximum cytokine release. Moreover, by optimizing the affinity of both parts of the molecule, it is possible to improve the selectivity. These results could be effectively modelled based on kinetic proofreading with limited signalling. This model explained the experimental observation that strong binding at both ends of the molecules leads to reduced activity, through very stable target-bispecific-effector complexes leading to CD3 entering a non-signalling dark state. These findings have important implications for the design of anti-CD3-based bispecifics with optimal biophysical parameters for both activity and specificity.
Keywords: T-cell activation; bispecific; cluster of differentiation 3; cytokine release; cytotoxicity.
© The Author(s) 2023. Published by Oxford University Press on behalf of the British Society for Immunology.
Conflict of interest statement
I.B.R., R.O., S.H., and P.B.K. are current employees of Immunocore and hold Immunocore stock and/or options.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

