ESYT1 tethers the ER to mitochondria and is required for mitochondrial lipid and calcium homeostasis
- PMID: 37931956
- PMCID: PMC10627786
- DOI: 10.26508/lsa.202302335
ESYT1 tethers the ER to mitochondria and is required for mitochondrial lipid and calcium homeostasis
Abstract
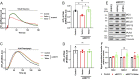
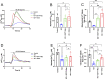
Mitochondria interact with the ER at structurally and functionally specialized membrane contact sites known as mitochondria-ER contact sites (MERCs). Combining proximity labelling (BioID), co-immunoprecipitation, confocal microscopy and subcellular fractionation, we found that the ER resident SMP-domain protein ESYT1 was enriched at MERCs, where it forms a complex with the outer mitochondrial membrane protein SYNJ2BP. BioID analyses using ER-targeted, outer mitochondrial membrane-targeted, and MERC-targeted baits, confirmed the presence of this complex at MERCs and the specificity of the interaction. Deletion of ESYT1 or SYNJ2BP reduced the number and length of MERCs. Loss of the ESYT1-SYNJ2BP complex impaired ER to mitochondria calcium flux and provoked a significant alteration of the mitochondrial lipidome, most prominently a reduction of cardiolipins and phosphatidylethanolamines. Both phenotypes were rescued by reexpression of WT ESYT1 and an artificial mitochondria-ER tether. Together, these results reveal a novel function for ESYT1 in mitochondrial and cellular homeostasis through its role in the regulation of MERCs.
© 2023 Janer et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

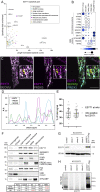

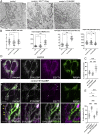
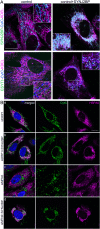
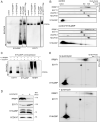




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
