Immunosuppressive low-density neutrophils in the blood of cancer patients display a mature phenotype
- PMID: 37931958
- PMCID: PMC10628041
- DOI: 10.26508/lsa.202302332
Immunosuppressive low-density neutrophils in the blood of cancer patients display a mature phenotype
Abstract
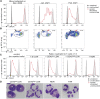
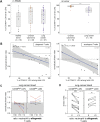
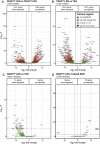
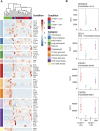
The presence of human neutrophils in the tumor microenvironment is strongly correlated to poor overall survival. Most previous studies have focused on the immunosuppressive capacities of low-density neutrophils (LDN), also referred to as granulocytic myeloid-derived suppressor cells, which are elevated in number in the blood of many cancer patients. We observed two types of LDN in the blood of lung cancer and ovarian carcinoma patients: CD45high LDN, which suppressed T-cell proliferation and displayed mature morphology, and CD45low LDN, which were immature and non-suppressive. We simultaneously evaluated the classical normal-density neutrophils (NDN) and, when available, tumor-associated neutrophils. We observed that NDN from cancer patients suppressed T-cell proliferation, and NDN from healthy donors did not, despite few transcriptomic differences. Hence, the immunosuppression mediated by neutrophils in the blood of cancer patients is not dependent on the cells' density but rather on their maturity.
© 2023 Vanhaver et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures










References
-
- Aarts CEM, Hiemstra IH, Béguin EP, Hoogendijk AJ, Bouchmal S, Van Houdt M, Tool ATJ, Mul E, Jansen MH, Janssen H, et al. (2019) Activated neutrophils exert myeloid-derived suppressor cell activity damaging T cells beyond repair. Blood Adv 3: 3562–3574. 10.1182/bloodadvances.2019031609 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
