FNIP1 abrogation promotes functional revascularization of ischemic skeletal muscle by driving macrophage recruitment
- PMID: 37932296
- PMCID: PMC10628247
- DOI: 10.1038/s41467-023-42690-9
FNIP1 abrogation promotes functional revascularization of ischemic skeletal muscle by driving macrophage recruitment
Abstract
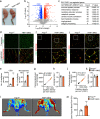
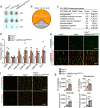
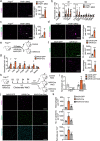
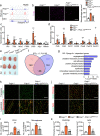
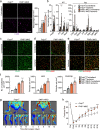
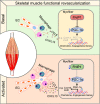
Ischaemia of the heart and limbs attributable to compromised blood supply is a major cause of mortality and morbidity. The mechanisms of functional angiogenesis remain poorly understood, however. Here we show that FNIP1 plays a critical role in controlling skeletal muscle functional angiogenesis, a process pivotal for muscle revascularization during ischemia. Muscle FNIP1 expression is down-regulated by exercise. Genetic overexpression of FNIP1 in myofiber causes limited angiogenesis in mice, whereas its myofiber-specific ablation markedly promotes the formation of functional blood vessels. Interestingly, the increased muscle angiogenesis is independent of AMPK but due to enhanced macrophage recruitment in FNIP1-depleted muscles. Mechanistically, myofiber FNIP1 deficiency induces PGC-1α to activate chemokine gene transcription, thereby driving macrophage recruitment and muscle angiogenesis program. Furthermore, in a mouse hindlimb ischemia model of peripheral artery disease, the loss of myofiber FNIP1 significantly improved the recovery of blood flow. Thus, these results reveal a pivotal role of FNIP1 as a negative regulator of functional angiogenesis in muscle, offering insight into potential therapeutic strategies for ischemic diseases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

