Genetically encoded multimeric tags for subcellular protein localization in cryo-EM
- PMID: 37932397
- PMCID: PMC10703698
- DOI: 10.1038/s41592-023-02053-0
Genetically encoded multimeric tags for subcellular protein localization in cryo-EM
Abstract
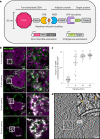
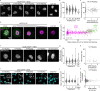
Cryo-electron tomography (cryo-ET) allows for label-free high-resolution imaging of macromolecular assemblies in their native cellular context. However, the localization of macromolecules of interest in tomographic volumes can be challenging. Here we present a ligand-inducible labeling strategy for intracellular proteins based on fluorescent, 25-nm-sized, genetically encoded multimeric particles (GEMs). The particles exhibit recognizable structural signatures, enabling their automated detection in cryo-ET data by convolutional neural networks. The coupling of GEMs to green fluorescent protein-tagged macromolecules of interest is triggered by addition of a small-molecule ligand, allowing for time-controlled labeling to minimize disturbance to native protein function. We demonstrate the applicability of GEMs for subcellular-level localization of endogenous and overexpressed proteins across different organelles in human cells using cryo-correlative fluorescence and cryo-ET imaging. We describe means for quantifying labeling specificity and efficiency, and for systematic optimization for rare and abundant protein targets, with emphasis on assessing the potential effects of labeling on protein function.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
- 419120233/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 402723784/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 101028297/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
- CDA00045/2019/Human Frontier Science Program (HFSP)
LinkOut - more resources
Full Text Sources
Research Materials

