CRISPR-Cas9-based functional interrogation of unconventional translatome reveals human cancer dependency on cryptic non-canonical open reading frames
- PMID: 37932451
- PMCID: PMC10716047
- DOI: 10.1038/s41594-023-01117-1
CRISPR-Cas9-based functional interrogation of unconventional translatome reveals human cancer dependency on cryptic non-canonical open reading frames
Abstract
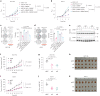
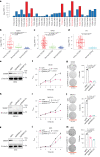
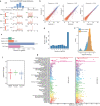
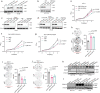
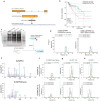
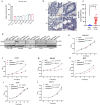
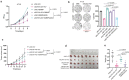
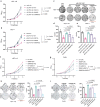
Emerging evidence suggests that cryptic translation beyond the annotated translatome produces proteins with developmental or physiological functions. However, functions of cryptic non-canonical open reading frames (ORFs) in cancer remain largely unknown. To fill this gap and systematically identify colorectal cancer (CRC) dependency on non-canonical ORFs, we apply an integrative multiomic strategy, combining ribosome profiling and a CRISPR-Cas9 knockout screen with large-scale analysis of molecular and clinical data. Many such ORFs are upregulated in CRC compared to normal tissues and are associated with clinically relevant molecular subtypes. We confirm the in vivo tumor-promoting function of the microprotein SMIMP, encoded by a primate-specific, long noncoding RNA, the expression of which is associated with poor prognosis in CRC, is low in normal tissues and is specifically elevated in CRC and several other cancer types. Mechanistically, SMIMP interacts with the ATPase-forming domains of SMC1A, the core subunit of the cohesin complex, and facilitates SMC1A binding to cis-regulatory elements to promote epigenetic repression of the tumor-suppressive cell cycle regulators encoded by CDKN1A and CDKN2B. Thus, our study reveals a cryptic microprotein as an important component of cohesin-mediated gene regulation and suggests that the 'dark' proteome, encoded by cryptic non-canonical ORFs, may contain potential therapeutic or diagnostic targets.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

