GATA transcription factors drive initial Xist upregulation after fertilization through direct activation of long-range enhancers
- PMID: 37932452
- PMCID: PMC10635832
- DOI: 10.1038/s41556-023-01266-x
GATA transcription factors drive initial Xist upregulation after fertilization through direct activation of long-range enhancers
Abstract
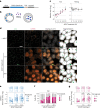
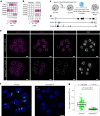
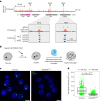
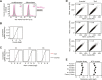
X-chromosome inactivation (XCI) balances gene expression between the sexes in female mammals. Shortly after fertilization, upregulation of Xist RNA from one X chromosome initiates XCI, leading to chromosome-wide gene silencing. XCI is maintained in all cell types, except the germ line and the pluripotent state where XCI is reversed. The mechanisms triggering Xist upregulation have remained elusive. Here we identify GATA transcription factors as potent activators of Xist. Through a pooled CRISPR activation screen in murine embryonic stem cells, we demonstrate that GATA1, as well as other GATA transcription factors can drive ectopic Xist expression. Moreover, we describe GATA-responsive regulatory elements in the Xist locus bound by different GATA factors. Finally, we show that GATA factors are essential for XCI induction in mouse preimplantation embryos. Deletion of GATA1/4/6 or GATA-responsive Xist enhancers in mouse zygotes effectively prevents Xist upregulation. We propose that the activity or complete absence of various GATA family members controls initial Xist upregulation, XCI maintenance in extra-embryonic lineages and XCI reversal in the epiblast.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. - PubMed
-
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. - PubMed
-
- Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. - PubMed
-
- Mak W, et al. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. - PubMed
-
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

