Alteration of chromatin high-order conformation associated with oxaliplatin resistance acquisition in colorectal cancer cells
- PMID: 37933235
- PMCID: PMC10624369
- DOI: 10.1002/EXP.20220136
Alteration of chromatin high-order conformation associated with oxaliplatin resistance acquisition in colorectal cancer cells
Abstract
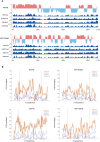
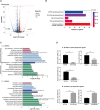
Oxaliplatin is a first-line chemotherapy drug widely adopted in colorectal cancer (CRC) treatment. However, a large proportion of patients tend to become resistant to oxaliplatin, causing chemotherapy to fail. At present, researches on oxaliplatin resistance mainly focus on the genetic and epigenetic alterations during cancer evolution, while the characteristics of high-order three-dimensional (3D) conformation of genome are yet to be explored. In order to investigate the chromatin conformation alteration during oxaliplatin resistance, we performed multi-omics study by combining DLO Hi-C, ChIP-seq as well as RNA-seq technologies on the established oxaliplatin-resistant cell line HCT116-OxR, as well as the control cell line HCT116. The results indicate that 19.33% of the genome regions have A/B compartments transformation after drug resistance, further analysis of the genes converted by A/B compartments reveals that the acquisition of oxaliplatin resistance in tumor cells is related to the reduction of reactive oxygen species and enhanced metastatic capacity. Our research reveals the spatial chromatin structural difference between CRC cells and oxaliplatin resistant cells based on the DLO Hi-C and other epigenetic omics experiments. More importantly, we provide potential targets for oxaliplatin-resistant cancer treatment and a new way to investigate drug resistance behavior under the perspective of 3D genome alteration.
Keywords: 3D spatial structure; colorectal cancer; multi‐omics; oxaliplatin resistance.
© 2023 The Authors. Exploration published by Henan University and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
