The ChEMBL Database in 2023: a drug discovery platform spanning multiple bioactivity data types and time periods
- PMID: 37933841
- PMCID: PMC10767899
- DOI: 10.1093/nar/gkad1004
The ChEMBL Database in 2023: a drug discovery platform spanning multiple bioactivity data types and time periods
Abstract
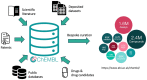
ChEMBL (https://www.ebi.ac.uk/chembl/) is a manually curated, high-quality, large-scale, open, FAIR and Global Core Biodata Resource of bioactive molecules with drug-like properties, previously described in the 2012, 2014, 2017 and 2019 Nucleic Acids Research Database Issues. Since its introduction in 2009, ChEMBL's content has changed dramatically in size and diversity of data types. Through incorporation of multiple new datasets from depositors since the 2019 update, ChEMBL now contains slightly more bioactivity data from deposited data vs data extracted from literature. In collaboration with the EUbOPEN consortium, chemical probe data is now regularly deposited into ChEMBL. Release 27 made curated data available for compounds screened for potential anti-SARS-CoV-2 activity from several large-scale drug repurposing screens. In addition, new patent bioactivity data have been added to the latest ChEMBL releases, and various new features have been incorporated, including a Natural Product likeness score, updated flags for Natural Products, a new flag for Chemical Probes, and the initial annotation of the action type for ∼270 000 bioactivity measurements.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

