DescribePROT in 2023: more, higher-quality and experimental annotations and improved data download options
- PMID: 37933852
- PMCID: PMC10767971
- DOI: 10.1093/nar/gkad985
DescribePROT in 2023: more, higher-quality and experimental annotations and improved data download options
Abstract
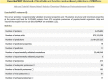
The DescribePROT database of amino acid-level descriptors of protein structures and functions was substantially expanded since its release in 2020. This expansion includes substantial increase in the size, scope, and quality of the underlying data, the addition of experimental structural information, the inclusion of new data download options, and an upgraded graphical interface. DescribePROT currently covers 19 structural and functional descriptors for proteins in 273 reference proteomes generated by 11 accurate and complementary predictive tools. Users can search our resource in multiple ways, interact with the data using the graphical interface, and download data at various scales including individual proteins, entire proteomes, and whole database. The annotations in DescribePROT are useful for a broad spectrum of studies that include investigations of protein structure and function, development and validation of predictive tools, and to support efforts in understanding molecular underpinnings of diseases and development of therapeutics. DescribePROT can be freely accessed at http://biomine.cs.vcu.edu/servers/DESCRIBEPROT/.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G.V., Christie C.H., Dalenberg K., Di Costanzo L., Duarte J.M.et al. .. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021; 49:D437–D451. - PMC - PubMed
-
- Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chao H., Chen L., Craig P.A., Crichlow G.V., Dalenberg K., Duarte J.M.et al. .. RCSB Protein Data Bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023; 51:D488–D508. - PMC - PubMed
-
- Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A.et al. .. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022; 50:D439–D444. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources