Refining PRRSV-2 genetic classification based on global ORF5 sequences and investigation of their geographic distributions and temporal changes
- PMID: 37933982
- PMCID: PMC10848785
- DOI: 10.1128/spectrum.02916-23
Refining PRRSV-2 genetic classification based on global ORF5 sequences and investigation of their geographic distributions and temporal changes
Abstract
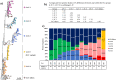
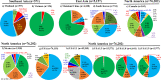
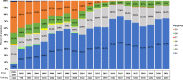
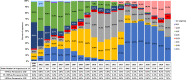
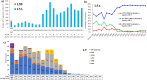
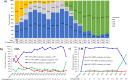
In this study, comprehensive analysis of 82,237 global porcine reproductive and respiratory syndrome virus type 2 (PRRSV-2) open reading frame 5 sequences spanning from 1989 to 2021 refined PRRSV-2 genetic classification system, which defines 11 lineages and 21 sublineages and provides flexibility for growth if additional lineages, sublineages, or more granular classifications are needed in the future. Geographic distribution and temporal changes of PRRSV-2 were investigated in detail. This is a thorough study describing the molecular epidemiology of global PRRSV-2. In addition, the reference sequences based on the refined genetic classification system are made available to the public for future epidemiological and diagnostic applications worldwide. The data from this study will facilitate global standardization and application of PRRSV-2 genetic classification.
Keywords: ORF5; PRRSV-2; genetic classification; lineage; molecular epidemiology; restriction fragment length polymorphism analysis; sublineage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Holtkamp D, Kliebenstein J, Neumann E, Zimmerman J, Rotto H, Yoder T, Wang C, Yeske P, Mowrer C, Haley C. 2013. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on U.S. pork producers. J Swine Health Prod 21:72–84.
-
- Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest 4:127–133. doi: 10.1177/104063879200400202 - DOI - PubMed
-
- Cavanagh D. 1997. Nidovirales: a new order comprising coronaviridae and arteriviridae. Arch Virol 142:629–633. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

