Development of simple, rapid, and sensitive methods for detection of hepatitis C virus RNA from whole blood using reverse transcription loop-mediated isothermal amplification
- PMID: 37933990
- PMCID: PMC10662345
- DOI: 10.1128/jcm.00771-23
Development of simple, rapid, and sensitive methods for detection of hepatitis C virus RNA from whole blood using reverse transcription loop-mediated isothermal amplification
Abstract
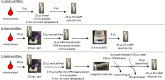
Hepatitis C virus (HCV) infection is an underdiagnosed global health problem. Diagnosis of current HCV infections typically requires testing for HCV RNA using high-complexity laboratory tests. Methods for the detection of HCV RNA that are simple, inexpensive, rapid, and compatible with use outside of a laboratory setting are very important in order to improve access to hepatitis C diagnostic testing and facilitate accelerated linkage to care. We developed and evaluated three simple workflows for extracting HCV RNA from small volumes of whole blood for use in a sensitive, pan-genotypic RT-LAMP assay. The water workflow uses osmotic stress to release HCV RNA and has a limit of detection of 4.3 log10(IU/mL) (95% CI 4.0-4.9). The heat workflow uses a heating step to release HCV RNA and has a limit of detection of 4.2 log10(IU/mL) (95% CI 3.8-5.1). The bead workflow, which uses chemical lysis of the sample and a streamlined paramagnetic solid phase reversible immobilization bead procedure for nucleic acid purification, has a limit of detection of 2.8 log10(IU/mL) (95% CI 2.5-3.4). When used to test whole blood spiked with HCV RNA-positive plasma samples in which most HCV levels were below 5.0 log10(IU/mL), the water, heat, and bead workflows detected HCV RNA in 69%, 75%, and 94% of samples, respectively. These workflows are compatible with visual lateral flow dipsticks, and each takes less than 60 min from sample to result. Each workflow can be performed with minimal and inexpensive equipment. With further procedural simplifications, these workflows may form the basis of assays for the point-of-care diagnosis of HCV infections.
Keywords: RNA detection; assay development; hepatitis C virus; point of care.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- World Health Organization . 2021. Global progress report on HIV, viral hepatitis and sexually transmitted infections, 2021: accountability for the global health sector strategies 2016–2021: actions for impact: web annex 2: data methods.
-
- Yin S, Barker L, White JZ, Jiles RB. 2019. Sofosbuvir-based regimens for chronic hepatitis C in a well-insured U.S. population: patient characteristics, treatment adherence, effectiveness, and health care costs, 2013-2015. J Manag Care Spec Pharm 25:195–210. doi: 10.18553/jmcp.2019.25.2.195 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

