CRISPR Screening Identifies Mechanisms of Resistance to KRASG12C and SHP2 Inhibitor Combinations in Non-Small Cell Lung Cancer
- PMID: 37934115
- PMCID: PMC10722132
- DOI: 10.1158/0008-5472.CAN-23-1127
CRISPR Screening Identifies Mechanisms of Resistance to KRASG12C and SHP2 Inhibitor Combinations in Non-Small Cell Lung Cancer
Abstract
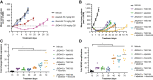
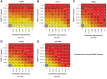
Although KRASG12C inhibitors show clinical activity in patients with KRAS G12C mutated non-small cell lung cancer (NSCLC) and other solid tumor malignancies, response is limited by multiple mechanisms of resistance. The KRASG12C inhibitor JDQ443 shows enhanced preclinical antitumor activity combined with the SHP2 inhibitor TNO155, and the combination is currently under clinical evaluation. To identify rational combination strategies that could help overcome or prevent some types of resistance, we evaluated the duration of tumor responses to JDQ443 ± TNO155, alone or combined with the PI3Kα inhibitor alpelisib and/or the cyclin-dependent kinase 4/6 inhibitor ribociclib, in xenograft models derived from a KRASG12C-mutant NSCLC line and investigated the genetic mechanisms associated with loss of response to combined KRASG12C/SHP2 inhibition. Tumor regression by single-agent JDQ443 at clinically relevant doses lasted on average 2 weeks and was increasingly extended by the double, triple, or quadruple combinations. Growth resumption was accompanied by progressively increased KRAS G12C amplification. Functional genome-wide CRISPR screening in KRASG12C-dependent NSCLC lines with distinct mutational profiles to identify adaptive mechanisms of resistance revealed sensitizing and rescuing genetic interactions with KRASG12C/SHP2 coinhibition; FGFR1 loss was the strongest sensitizer, and PTEN loss the strongest rescuer. Consistently, the antiproliferative activity of KRASG12C/SHP2 inhibition was strongly enhanced by PI3K inhibitors. Overall, KRAS G12C amplification and alterations of the MAPK/PI3K pathway were predominant mechanisms of resistance to combined KRASG12C/SHP2 inhibitors in preclinical settings. The biological nodes identified by CRISPR screening might provide additional starting points for effective combination treatments.
Significance: Identification of resistance mechanisms to KRASG12C/SHP2 coinhibition highlights the need for additional combination therapies for lung cancer beyond on-pathway combinations and offers the basis for development of more effective combination approaches. See related commentary by Johnson and Haigis, p. 4005.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures




Comment in
-
All Roads Lead to Rome: YAP/TAZ Activity Influences Efficacy of KRASG12C Inhibitors.Cancer Res. 2023 Dec 15;83(24):4005-4007. doi: 10.1158/0008-5472.CAN-23-3547. Cancer Res. 2023. PMID: 38098448
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

