Durable responses to ATR inhibition with ceralasertib in tumors with genomic defects and high inflammation
- PMID: 37934611
- PMCID: PMC10786692
- DOI: 10.1172/JCI175369
Durable responses to ATR inhibition with ceralasertib in tumors with genomic defects and high inflammation
Abstract
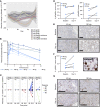
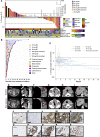
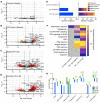
BACKGROUNDPhase 1 study of ATRinhibition alone or with radiation therapy (PATRIOT) was a first-in-human phase I study of the oral ATR (ataxia telangiectasia and Rad3-related) inhibitor ceralasertib (AZD6738) in advanced solid tumors.METHODSThe primary objective was safety. Secondary objectives included assessment of antitumor responses and pharmacokinetic (PK) and pharmacodynamic (PD) studies. Sixty-seven patients received 20-240 mg ceralasertib BD continuously or intermittently (14 of a 28-day cycle).RESULTSIntermittent dosing was better tolerated than continuous, which was associated with dose-limiting hematological toxicity. The recommended phase 2 dose of ceralasertib was 160 mg twice daily for 2 weeks in a 4-weekly cycle. Modulation of target and increased DNA damage were identified in tumor and surrogate PD. There were 5 (8%) confirmed partial responses (PRs) (40-240 mg BD), 34 (52%) stable disease (SD), including 1 unconfirmed PR, and 27 (41%) progressive disease. Durable responses were seen in tumors with loss of AT-rich interactive domain-containing protein 1A (ARID1A) and DNA damage-response defects. Treatment-modulated tumor and systemic immune markers and responding tumors were more immune inflamed than nonresponding.CONCLUSIONCeralasertib monotherapy was tolerated at 160 mg BD intermittently and associated with antitumor activity.TRIAL REGISTRATIONClinicaltrials.gov: NCT02223923, EudraCT: 2013-003994-84.FUNDINGCancer Research UK, AstraZeneca, UK Department of Health (National Institute for Health Research), Rosetrees Trust, Experimental Cancer Medicine Centre.
Keywords: Cancer immunotherapy; DNA repair; Drug therapy; Oncology.
Figures



References
-
- Lau A, et al. Abstract C60: Pre-clinical efficacy of the ATR inhibitor AZD6738 in combination with the PARP inhibitor olaparib. Mol Cancer Ther. 2015;14(12 suppl 2):C60. doi: 10.1158/1535-7163.TARG-15-C60. - DOI
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

