CGG toolkit: Software components for computational genomics
- PMID: 37934729
- PMCID: PMC10629618
- DOI: 10.1371/journal.pcbi.1011498
CGG toolkit: Software components for computational genomics
Abstract
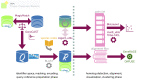
Public-domain availability for bioinformatics software resources is a key requirement that ensures long-term permanence and methodological reproducibility for research and development across the life sciences. These issues are particularly critical for widely used, efficient, and well-proven methods, especially those developed in research settings that often face funding discontinuities. We re-launch a range of established software components for computational genomics, as legacy version 1.0.1, suitable for sequence matching, masking, searching, clustering and visualization for protein family discovery, annotation and functional characterization on a genome scale. These applications are made available online as open source and include MagicMatch, GeneCAST, support scripts for CoGenT-like sequence collections, GeneRAGE and DifFuse, supported by centrally administered bioinformatics infrastructure funding. The toolkit may also be conceived as a flexible genome comparison software pipeline that supports research in this domain. We illustrate basic use by examples and pictorial representations of the registered tools, which are further described with appropriate documentation files in the corresponding GitHub release.
Copyright: © 2023 Vasileiou et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Hinchliff CE, Smith SA, Allman JF, Burleigh JG, Chaudhary R, Coghill LM, et al.. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc Natl Acad Sci U S A. 2015;112(41):12764–9. Epub 20150918. doi: 10.1073/pnas.1423041112 ; PubMed Central PMCID: PMC4611642. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

