KSHV vIL-6 enhances inflammatory responses by epigenetic reprogramming
- PMID: 37934757
- PMCID: PMC10656005
- DOI: 10.1371/journal.ppat.1011771
KSHV vIL-6 enhances inflammatory responses by epigenetic reprogramming
Abstract
Kaposi sarcoma-associated herpesvirus (KSHV) inflammatory cytokine syndrome (KICS) is a newly described chronic inflammatory disease condition caused by KSHV infection and is characterized by high KSHV viral load and sustained elevations of serum KSHV-encoded IL-6 (vIL-6) and human IL-6 (hIL-6). KICS has significant immortality and greater risks of other complications, including malignancies. Although prolonged inflammatory vIL-6 exposure by persistent KSHV infection is expected to have key roles in subsequent disease development, the biological effects of prolonged vIL-6 exposure remain elusive. Using thiol(SH)-linked alkylation for the metabolic (SLAM) sequencing and Cleavage Under Target & Release Using Nuclease analysis (CUT&RUN), we studied the effect of prolonged vIL-6 exposure in chromatin landscape and resulting cytokine production. The studies showed that prolonged vIL-6 exposure increased Bromodomain containing 4 (BRD4) and histone H3 lysine 27 acetylation co-occupancies on chromatin, and the recruitment sites were frequently co-localized with poised RNA polymerase II with associated enzymes. Increased BRD4 recruitment on promoters was associated with increased and prolonged NF-κB p65 binding after the lipopolysaccharide stimulation. The p65 binding resulted in quicker and sustained transcription bursts from the promoters; this mechanism increased total amounts of hIL-6 and IL-10 in tissue culture. Pretreatment with the BRD4 inhibitors, OTX015 and MZ1, eliminated the enhanced inflammatory cytokine production. These findings suggest that persistent vIL-6 exposure may establish a chromatin landscape favorable for the reactivation of inflammatory responses in monocytes. This epigenetic memory may explain the greater risk of chronic inflammatory disease development in KSHV-infected individuals.
Copyright: © 2023 Inagaki et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: M.S. and Y.I. are founders of VGN BIO, Inc. and have a pending patent application for the use of vIL-6 for therapeutic purposes. No other authors have financial interests.
Figures


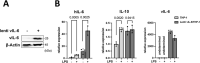


Update of
-
KSHV vIL-6 Enhances Inflammatory Responses by Epigenetic Reprogramming.bioRxiv [Preprint]. 2023 Jul 23:2023.06.25.546454. doi: 10.1101/2023.06.25.546454. bioRxiv. 2023. Update in: PLoS Pathog. 2023 Nov 7;19(11):e1011771. doi: 10.1371/journal.ppat.1011771. PMID: 37503036 Free PMC article. Updated. Preprint.
Similar articles
-
KSHV vIL-6 Enhances Inflammatory Responses by Epigenetic Reprogramming.bioRxiv [Preprint]. 2023 Jul 23:2023.06.25.546454. doi: 10.1101/2023.06.25.546454. bioRxiv. 2023. Update in: PLoS Pathog. 2023 Nov 7;19(11):e1011771. doi: 10.1371/journal.ppat.1011771. PMID: 37503036 Free PMC article. Updated. Preprint.
-
Kaposi's Sarcoma-Associated Herpesvirus Viral Interleukin-6 Signaling Upregulates Integrin β3 Levels and Is Dependent on STAT3.J Virol. 2020 Feb 14;94(5):e01384-19. doi: 10.1128/JVI.01384-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31801855 Free PMC article.
-
Virally encoded interleukin-6 facilitates KSHV replication in monocytes and induction of dysfunctional macrophages.PLoS Pathog. 2023 Oct 26;19(10):e1011703. doi: 10.1371/journal.ppat.1011703. eCollection 2023 Oct. PLoS Pathog. 2023. PMID: 37883374 Free PMC article.
-
KSHV-associated multicentric Castleman disease: A tangle of different entities requiring multitarget treatment strategies.Int J Cancer. 2015 Jul 15;137(2):251-61. doi: 10.1002/ijc.28923. Epub 2014 May 5. Int J Cancer. 2015. PMID: 24771491 Review.
-
Pathological Features of Kaposi's Sarcoma-Associated Herpesvirus Infection.Adv Exp Med Biol. 2018;1045:357-376. doi: 10.1007/978-981-10-7230-7_16. Adv Exp Med Biol. 2018. PMID: 29896675 Review.
Cited by
-
Studies on Gene Enhancer with KSHV mini-chromatin.bioRxiv [Preprint]. 2025 Mar 24:2025.03.24.644916. doi: 10.1101/2025.03.24.644916. bioRxiv. 2025. PMID: 40196677 Free PMC article. Preprint.
-
Viral oncogenesis in cancer: from mechanisms to therapeutics.Signal Transduct Target Ther. 2025 May 12;10(1):151. doi: 10.1038/s41392-025-02197-9. Signal Transduct Target Ther. 2025. PMID: 40350456 Free PMC article. Review.
-
The Role of vIL-6 in KSHV-Mediated Immune Evasion and Tumorigenesis.Viruses. 2024 Dec 10;16(12):1900. doi: 10.3390/v16121900. Viruses. 2024. PMID: 39772207 Free PMC article. Review.
-
Divergent Processing of Cell Stress Signals as the Basis of Cancer Progression: Licensing NFκB on Chromatin.Int J Mol Sci. 2024 Aug 7;25(16):8621. doi: 10.3390/ijms25168621. Int J Mol Sci. 2024. PMID: 39201306 Free PMC article. Review.
-
HIV-associated cancers and lymphoproliferative disorders caused by Kaposi sarcoma herpesvirus and Epstein-Barr virus.Clin Microbiol Rev. 2024 Sep 12;37(3):e0002223. doi: 10.1128/cmr.00022-23. Epub 2024 Jun 20. Clin Microbiol Rev. 2024. PMID: 38899877 Free PMC article. Review.
References
-
- Mizobuchi H, Yamamoto K, Tsutsui S, Yamashita M, Nakata Y, Inagawa H, et al. A unique hybrid characteristic having both pro- and anti-inflammatory phenotype transformed by repetitive low-dose lipopolysaccharide in C8-B4 microglia. Sci Rep. 2020;10(1):8945. Epub 20200602. doi: 10.1038/s41598-020-65998-8 ; PubMed Central PMCID: PMC7265460. - DOI - PMC - PubMed
-
- Morein D, Rubinstein-Achiasaf L, Brayer H, Dorot O, Pichinuk E, Ben-Yaakov H, et al. Continuous Inflammatory Stimulation Leads via Metabolic Plasticity to a Prometastatic Phenotype in Triple-Negative Breast Cancer Cells. Cells. 2021;10(6). Epub 20210531. doi: 10.3390/cells10061356 ; PubMed Central PMCID: PMC8229065. - DOI - PMC - PubMed

