Tumor and immune cell types interact to produce heterogeneous phenotypes of pediatric high-grade glioma
- PMID: 37934854
- PMCID: PMC10912009
- DOI: 10.1093/neuonc/noad207
Tumor and immune cell types interact to produce heterogeneous phenotypes of pediatric high-grade glioma
Abstract
Background: Pediatric high-grade gliomas (PHGG) are aggressive brain tumors with 5-year survival rates ranging from <2% to 20% depending upon subtype. PHGG presents differently from patient to patient and is intratumorally heterogeneous, posing challenges in designing therapies. We hypothesized that heterogeneity occurs because PHGG comprises multiple distinct tumor and immune cell types in varying proportions, each of which may influence tumor characteristics.
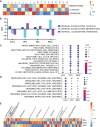
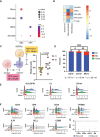
Methods: We obtained 19 PHGG samples from our institution's pediatric brain tumor bank. We constructed a comprehensive transcriptomic dataset at the single-cell level using single-cell RNA-Seq (scRNA-Seq), identified known glial and immune cell types, and performed differential gene expression and gene set enrichment analysis. We conducted multi-channel immunofluorescence (IF) staining to confirm the transcriptomic results.
Results: Our PHGG samples included 3 principal predicted tumor cell types: astrocytes, oligodendrocyte progenitors (OPCs), and mesenchymal-like cells (Mes). These cell types differed in their gene expression profiles, pathway enrichment, and mesenchymal character. We identified a macrophage population enriched in mesenchymal and inflammatory gene expression as a possible source of mesenchymal tumor characteristics. We found evidence of T-cell exhaustion and suppression.
Conclusions: PHGG comprises multiple distinct proliferating tumor cell types. Microglia-derived macrophages may drive mesenchymal gene expression in PHGG. The predicted Mes tumor cell population likely derives from OPCs. The variable tumor cell populations rely on different oncogenic pathways and are thus likely to vary in their responses to therapy.
Keywords: glioma; pediatric | scRNA-Seq.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Fangusaro J. Pediatric high-grade gliomas and diffuse intrinsic pontine gliomas. J Child Neurol. 2009;24(11):1409–1417. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

