The neuropeptidergic connectome of C. elegans
- PMID: 37935195
- PMCID: PMC7615469
- DOI: 10.1016/j.neuron.2023.09.043
The neuropeptidergic connectome of C. elegans
Abstract
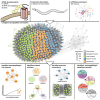
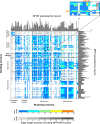
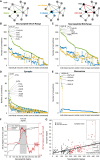
Efforts are ongoing to map synaptic wiring diagrams, or connectomes, to understand the neural basis of brain function. However, chemical synapses represent only one type of functionally important neuronal connection; in particular, extrasynaptic, "wireless" signaling by neuropeptides is widespread and plays essential roles in all nervous systems. By integrating single-cell anatomical and gene-expression datasets with biochemical analysis of receptor-ligand interactions, we have generated a draft connectome of neuropeptide signaling in the C. elegans nervous system. This network is characterized by high connection density, extended signaling cascades, autocrine foci, and a decentralized topology, with a large, highly interconnected core containing three constituent communities sharing similar patterns of input connectivity. Intriguingly, several key network hubs are little-studied neurons that appear specialized for peptidergic neuromodulation. We anticipate that the C. elegans neuropeptidergic connectome will serve as a prototype to understand how networks of neuromodulatory signaling are organized.
Keywords: C. elegans; connectomics; networks; neuromodulation; neuropeptides.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
-
- Csaba V., Sanja J., Martin G., Réza S., Nobuo U., James David B., Sara M., Konrad H., Luis Alberto B.-C., Elizabeth W., et al. Whole-animal connectome and cell-type complement of the three-segmented Platynereis dumerilii larva. bioRxiv. 2020 doi: 10.1101/2020.08.21.260984. - DOI
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

