SGF29 nuclear condensates reinforce cellular aging
- PMID: 37935676
- PMCID: PMC10630320
- DOI: 10.1038/s41421-023-00602-7
SGF29 nuclear condensates reinforce cellular aging
Erratum in
-
Publisher Correction: SGF29 nuclear condensates reinforce cellular aging.Cell Discov. 2024 May 30;10(1):61. doi: 10.1038/s41421-024-00690-z. Cell Discov. 2024. PMID: 38816368 Free PMC article. No abstract available.
-
Author Correction: SGF29 nuclear condensates reinforce cellular aging.Cell Discov. 2025 Feb 15;11(1):17. doi: 10.1038/s41421-025-00773-5. Cell Discov. 2025. PMID: 39955282 Free PMC article. No abstract available.
Abstract
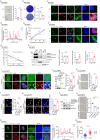
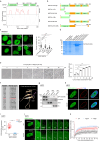
Phase separation, a biophysical segregation of subcellular milieus referred as condensates, is known to regulate transcription, but its impacts on physiological processes are less clear. Here, we demonstrate the formation of liquid-like nuclear condensates by SGF29, a component of the SAGA transcriptional coactivator complex, during cellular senescence in human mesenchymal progenitor cells (hMPCs) and fibroblasts. The Arg 207 within the intrinsically disordered region is identified as the key amino acid residue for SGF29 to form phase separation. Through epigenomic and transcriptomic analysis, our data indicated that both condensate formation and H3K4me3 binding of SGF29 are essential for establishing its precise chromatin location, recruiting transcriptional factors and co-activators to target specific genomic loci, and initiating the expression of genes associated with senescence, such as CDKN1A. The formation of SGF29 condensates alone, however, may not be sufficient to drive H3K4me3 binding or achieve transactivation functions. Our study establishes a link between phase separation and aging regulation, highlighting nuclear condensates as a functional unit that facilitate shaping transcriptional landscapes in aging.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
LinkOut - more resources
Full Text Sources

