NR5A2 connects zygotic genome activation to the first lineage segregation in totipotent embryos
- PMID: 37935903
- PMCID: PMC10709309
- DOI: 10.1038/s41422-023-00887-z
NR5A2 connects zygotic genome activation to the first lineage segregation in totipotent embryos
Abstract
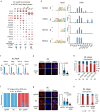
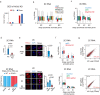
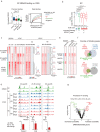
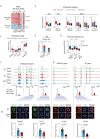
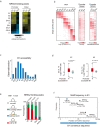
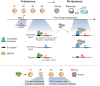
Zygotic genome activation (ZGA) marks the beginning of the embryonic program for a totipotent embryo, which gives rise to the inner cell mass (ICM) where pluripotent epiblast arises, and extraembryonic trophectoderm. However, how ZGA is connected to the first lineage segregation in mammalian embryos remains elusive. Here, we investigated the role of nuclear receptor (NR) transcription factors (TFs), whose motifs are highly enriched and accessible from the 2-cell (2C) to 8-cell (8C) stages in mouse embryos. We found that NR5A2, an NR TF strongly induced upon ZGA, was required for this connection. Upon Nr5a2 knockdown or knockout, embryos developed beyond 2C normally with the zygotic genome largely activated. However, 4-8C-specific gene activation was substantially impaired and Nr5a2-deficient embryos subsequently arrested at the morula stage. Genome-wide chromatin binding analysis showed that NR5A2-bound cis-regulatory elements in both 2C and 8C embryos are strongly enriched for B1 elements where its binding motif is embedded. NR5A2 was not required for the global opening of its binding sites in 2C embryos but was essential to the opening of its 8C-specific binding sites. These 8C-specific, but not 2C-specific, binding sites are enriched near genes involved in blastocyst and stem cell regulation, and are often bound by master pluripotency TFs in blastocysts and embryonic stem cells (ESCs). Importantly, NR5A2 regulated key pluripotency genes Nanog and Pou5f1/Oct4, and primitive endoderm regulatory genes including Gata6 among many early ICM genes, as well as key trophectoderm regulatory genes including Tead4 and Gata3 at the 8C stage. By contrast, master pluripotency TFs NANOG, SOX2, and OCT4 targeted both early and late ICM genes in mouse ESCs. Taken together, these data identify NR5A2 as a key regulator in totipotent embryos that bridges ZGA to the first lineage segregation during mouse early development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Lineage segregation in human pre-implantation embryos is specified by YAP1 and TEAD1.Hum Reprod. 2023 Aug 1;38(8):1484-1498. doi: 10.1093/humrep/dead107. Hum Reprod. 2023. PMID: 37295962
-
Lineage regulators TFAP2C and NR5A2 function as bipotency activators in totipotent embryos.Nat Struct Mol Biol. 2024 Jun;31(6):950-963. doi: 10.1038/s41594-023-01199-x. Epub 2024 Jan 19. Nat Struct Mol Biol. 2024. PMID: 38243114
-
TEAD4 regulates trophectoderm differentiation upstream of CDX2 in a GATA3-independent manner in the human preimplantation embryo.Hum Reprod. 2022 Jul 30;37(8):1760-1773. doi: 10.1093/humrep/deac138. Hum Reprod. 2022. PMID: 35700449
-
Genome folding and zygotic genome activation in mammalian preimplantation embryos.Curr Opin Genet Dev. 2024 Dec;89:102268. doi: 10.1016/j.gde.2024.102268. Epub 2024 Oct 9. Curr Opin Genet Dev. 2024. PMID: 39383545 Review.
-
Totipotency and lineage segregation in the human embryo.Mol Hum Reprod. 2014 Jul;20(7):599-618. doi: 10.1093/molehr/gau027. Epub 2014 Apr 3. Mol Hum Reprod. 2014. PMID: 24699365 Review.
Cited by
-
Mapping putative enhancers in mouse oocytes and early embryos reveals TCF3/12 as key folliculogenesis regulators.Nat Cell Biol. 2024 Jun;26(6):962-974. doi: 10.1038/s41556-024-01422-x. Epub 2024 Jun 5. Nat Cell Biol. 2024. PMID: 38839978
-
New Insights and Implications of Cell-Cell Interactions in Developmental Biology.Int J Mol Sci. 2025 Apr 23;26(9):3997. doi: 10.3390/ijms26093997. Int J Mol Sci. 2025. PMID: 40362237 Free PMC article. Review.
-
The transcription factor GABPA is a master regulator of naive pluripotency.Nat Cell Biol. 2025 Jan;27(1):48-58. doi: 10.1038/s41556-024-01554-0. Epub 2025 Jan 2. Nat Cell Biol. 2025. PMID: 39747581 Free PMC article.
-
A long noncoding RNA with enhancer-like function in pig zygotic genome activation.J Mol Cell Biol. 2025 Jun 28;17(1):mjae061. doi: 10.1093/jmcb/mjae061. J Mol Cell Biol. 2025. PMID: 39749834 Free PMC article.
-
ImputeHiFI: An Imputation Method for Multiplexed DNA FISH Data by Utilizing Single-Cell Hi-C and RNA FISH Data.Adv Sci (Weinh). 2024 Nov;11(42):e2406364. doi: 10.1002/advs.202406364. Epub 2024 Sep 12. Adv Sci (Weinh). 2024. PMID: 39264290 Free PMC article.
References
-
- Rossant J. Postimplantation development of blastomeres isolated from 4- and 8-cell mouse eggs. J. Embryol. Exp. Morphol. 1976;36:283–290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

