Inflammation-induced TRIM21 represses hepatic steatosis by promoting the ubiquitination of lipogenic regulators
- PMID: 37937648
- PMCID: PMC10721265
- DOI: 10.1172/jci.insight.164694
Inflammation-induced TRIM21 represses hepatic steatosis by promoting the ubiquitination of lipogenic regulators
Abstract
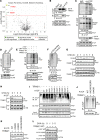
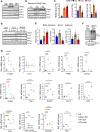
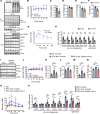
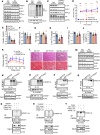
Nonalcoholic steatohepatitis (NASH) is a leading cause for chronic liver diseases. Current therapeutic options are limited due to an incomplete mechanistic understanding of how steatosis transitions to NASH. Here we show that the TRIM21 E3 ubiquitin ligase is induced by the synergistic actions of proinflammatory TNF-α and fatty acids in livers of humans and mice with NASH. TRIM21 ubiquitinates and degrades ChREBP, SREBP1, ACC1, and FASN, key regulators of de novo lipogenesis, and A1CF, an alternative splicing regulator of the high-activity ketohexokinase-C (KHK-C) isoform and rate-limiting enzyme of fructose metabolism. TRIM21-mediated degradation of these lipogenic activators improved steatosis and hyperglycemia as well as fructose and glucose tolerance. Our study identifies TRIM21 as a negative regulator of liver steatosis in NASH and provides mechanistic insights into an immunometabolic crosstalk that limits fatty acid synthesis and fructose metabolism during metabolic stress. Thus, enhancing this natural counteracting force of steatosis through inhibition of key lipogenic activators via TRIM21-mediated ubiquitination may provide a therapeutic opportunity to treat NASH.
Keywords: Diabetes; Hepatology.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

