Interactions between BRD4S, LOXL2, and MED1 drive cell cycle transcription in triple-negative breast cancer
- PMID: 37937685
- PMCID: PMC10701626
- DOI: 10.15252/emmm.202318459
Interactions between BRD4S, LOXL2, and MED1 drive cell cycle transcription in triple-negative breast cancer
Abstract
Triple-negative breast cancer (TNBC) often develops resistance to single-agent treatment, which can be circumvented using targeted combinatorial approaches. Here, we demonstrate that the simultaneous inhibition of LOXL2 and BRD4 synergistically limits TNBC proliferation in vitro and in vivo. Mechanistically, LOXL2 interacts in the nucleus with the short isoform of BRD4 (BRD4S), MED1, and the cell cycle transcriptional regulator B-MyB. These interactions sustain the formation of BRD4 and MED1 nuclear transcriptional foci and control cell cycle progression at the gene expression level. The pharmacological co-inhibition of LOXL2 and BRD4 reduces BRD4 nuclear foci, BRD4-MED1 colocalization, and the transcription of cell cycle genes, thus suppressing TNBC cell proliferation. Targeting the interaction between BRD4S and LOXL2 could be a starting point for the development of new anticancer strategies for the treatment of TNBC.
Keywords: cell cycle; combinatorial therapy; gene expression; triple-negative breast cancer.
© 2023 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
TVT and SP received funding from Pharmaxis.
Figures

Cell viability of high and low LOXL2‐expressing CCLE cell lines (mRNA levels) treated with different chemotherapeutic agents and BETi small molecules at the highest concentration (i.e., 10 μM). Color gradient indicates cell viability, with 8 being the highest and 0 the lowest. Significance was determined using an unpaired Student's t‐test with BH correction.
Analysis of the CPTAC proteomics dataset showing the protein abundance of BRD4 and LOXL2 in different breast cancer subtypes from tumor samples classified by subtype using vimentin (VIM), human epidermal growth factor receptor 2 (HER2), progesterone receptor (PGR), and estrogen receptor 1 (ESR1) protein abundance. N = 96 tumor samples. The significance of each cancer subtype against adjacent tissue was calculated using one‐way ANOVA with Tukey's post hoc test. The bottom and top fractions in the boxes represent the first and third quartiles, and the line, the median. Whiskers denote the interval between 1.5 times the interquartile range (IQR) and the median. Data beyond the end of the whiskers are plotted as outliers.
Representative Western blot analysis showing BRD4 and LOXL2 protein levels in three different TNBC cell lines. Tubulin is the loading control (ns: non‐specific). Three biological replicates were performed.
Cell viability assay of MDA‐MB‐231 cells infected with C or LOXL2 KD and treated with either DMSO or 2.5 μM of JQ1 for 24 and 48 h. Data were analyzed with MTT assay and normalized to condition C treated with DMSO. Data are shown as the mean of three independent biological replicates. The standard deviation is shown as error bars. Significance was determined using one‐way ANOVA multiple comparisons with Tukey's correction test.

BRD4 pulldown in MDA‐MB‐231 cells using irrelevant IgG as a negative control. Precipitates were analyzed by Western blot with the indicated antibodies. Three biological replicates were performed; ns: non‐specific.
Flag pulldown performed in MDA‐MB‐231 cells overexpressing either empty vector (EV), Flag‐tagged LOXL2 wild‐type (LOXL2wt), or the catalytically dead form of LOXL2 (LOXL2m). Precipitates were analyzed by Western blot with the indicated antibodies. Three biological replicates were performed. ns: non‐specific.
Schematic representation of BRD4‐GFP constructs used in (D).
Flag pulldown of HEK293T cells overexpressing either empty vector (EV) or a combination of the indicated constructs. Precipitates were analyzed by Western blot with the indicated antibodies. Three biological replicates were performed.
Details of docking models 4uyd_complex_3 (BD1), 2ouo_complex_5 (BD2), and their Asp→Phe mutant versions. The panel shows the superposition of LOXL2 (gray) docked to BD1 (cyan) and BD2 (green), with asparagines N140 and N433 mutated to phenylalanine (blue and yellow, respectively), both of which face the buried tryptophan W493 from LOXL2. On the left, zoomed visions of the mutants are shown superposed over their wild‐type structures.
Superimposition of selected docking models of BD1 (PDB: 3MXF) or BD2 (PDB: 3ONI) captured as crystallographic structures binding JQ1. Superimposition on 3MXF of the docking model of BD1 (4uyd_zdock_10; panel 1) or of BD2 (2ouo_zdock_4, panel 3) shows the compatibility of binding despite the presence of JQ1 in the AcK binding pocket. In contrast, superimposition on 3ONI of the docking model of BD1 (4uyd_zdock_3; panel 2) or of BD2 (2ouo_zdock_5, panel 4) shows the incompatibility of binding due to binding site competition of JQ1 and LOXL2.
BRD4 pulldown in MDA‐MB‐231 cells treated either with DMSO or with 5 μM of JQ1 for 24 h. IgGs were used as a negative control, and the precipitates were analyzed by Western blot with the indicated antibodies. Three biological replicates were performed. ns: non‐specific.
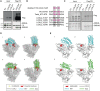
Flag pulldown in HEK293T cells overexpressing the empty vector (EV), LOXL2‐Flag wild‐type (LOXL2wt), or the catalytically dead form of LOXL2‐Flag (LOXL2m). Precipitates were analyzed by Western blot with the indicated antibodies. Three biological replicates were performed. ns: non‐specific.
Schematic representation of the double‐K H4‐mimic motif partially shared between H4, Twist, and LOXL2.
Flag pulldown of MDA‐MB‐231 cells overexpressing EV, LOXL2wt, LOXL2 R (the K209 residue mutated to R), or LOXL2 Q (the K209/K212 residues mutated to Q). Precipitates were analyzed by Western blot with the indicated antibodies. Three biological replicates were performed.
Selected docking models, two for each of the BDs. The docking poses 1 and 2 show the BD1 structure docked on LOXL2, corresponding to models 4uyd_zdock_10 and 4uyd_zdock_3, respectively (Tables EV2 and EV3), while 3 and 4 show the BD2 structures docked into LOXL2, corresponding to models 2ouo_zdock_4 and 2ouo_zdock_5, respectively (Tables EV2 and EV3). Red indicates the atomic representation of residues predicted to be fundamental for the modeled interaction (Table EV3), while yellow indicates the asparagines (N) N140 (BD1), and N433 (BD2). The molecular volumes of the four models are shown in light gray.
View of the four selected models highlighting LOXL2 histidines H626 and H628 whose mutations to glutamine did not affect the LOXL2 binding to BRD4. Panels 1 to 4 correspond to docking models 4uyd_complex_10, 4uyd_complex_3, 2ouo_complex_4, and 2ouo_complex_5, respectively (Table EV3).

Representative Western blot analysis of MDA‐MB‐231 cells infected with C or LOXL2 KD showing LOXL2 levels. Tubulin was used as a loading control. Three biological replicates were performed.
ATAC‐seq normalized coverage in MDA‐MB‐231 cells transduced with either C or LOXL2 KD, represented as the distance from the center of all peaks (bp = base pair).
Volcano Plot representation of the differential expression of genes between C and KD conditions. Significance was calculated using the Wald test with Benjamini–Hochberg correction used for multiple testing. Genes with adjusted P‐values <0.05 and abs (FC) > 1.5 were considered significant.
RNA‐seq logFC for genes associated with ATAC‐seq peaks in C and KD conditions which fall in promoter regions. The bottom and top fractions in the boxes represent the first and third quartiles, and the line, the median. Whiskers denote the interval between 1.5 times the interquartile range (IQR) and the median. Data beyond the end of the whiskers are plotted as outliers.
Gene Set Enrichment Analysis (GSEA) of the genes upregulated upon LOXL2 KD in the RNA‐seq dataset. Significance was calculated using a permutation test with Benjamini‐Hochberg correction used for multiple testing.
Number of BRD4 ChIP‐seq total or promoter peaks using Ab1 or Ab2 antibodies identified with MACS2.
Top‐10 GO‐terms identified either with Ab1 or Ab2 when analyzing promoter peaks with the mSigDB. Gene ratios and adjusted P‐values are reported on the left side of each GS. GSs shared among the top 10 of Ab1 and Ab2 are indicated in bold.
Representative Western blot analysis showing BRD4 levels in MDA‐MB‐231 cells infected with shControl), shBRD4 Long (BRD4L KD), and shBRD4 Short (BRD4S KD) isoforms. Tubulin was used as a loading control. Three biological replicates were performed. ns: non‐specific.
Heatmap of ChIP‐seq normalized signal (reads per genomic content) in all peaks in LOXL2 KD or control cells for the antibodies Ab1 and Ab2. The normalized signal is calculated for a region of −1 to 1 kb from the center of the peaks.
Normalized ATAC‐seq signal (reads per genomic content) in the ChIP‐seq peaks for Ab1 and Ab2 in LOXL2 KD or control cells. Significance was calculated using a two‐sample Kolmogorov–Smirnov test. The bottom and top fractions in the boxes represent the first and third quartiles, and the line, the median. Whiskers denote the interval between 1.5 times the interquartile range (IQR) and the median. Data beyond the end of the whiskers are plotted as outliers.
RNA‐seq logFC for genes associated with the peaks of Ab1 and Ab2, which fall in promoter regions in control or LOXL2 KD cells, respectively. Significance was calculated using a two‐sample Kolmogorov–Smirnov test. The bottom and top fractions in the boxes represent the first and third quartiles, and the line, the median. Whiskers denote the interval between 1.5 times the interquartile range (IQR) and the median. Data beyond the end of the whiskers are plotted as outliers.
Venn diagram showing the number of promoter genes identified with the ChIP‐seq with either Ab1 or Ab2 antibodies in control (left) or LOXL2 KD (right) cells. DREAM target genes identified in each condition are depicted in colored dots. The numbers on top of the dots represent the total number of promoters retrieved for each condition. The numbers below the dots are respectively the (upper) total number of DREAM target gene promoters retrieved in each condition and (lower) the condition‐relative percentage of DREAM target gene promoters identified (DREAM target gene promoters relative to all promoters).
Venn diagram showing the overlap between the total number of peaks detected with the ChIP‐seq with either Ab1 or Ab2 in control (left) or LOXL2 KD (right) conditions. The overlap of Ab1 and Ab2 peaks is shown as a percentage (intersection relative to Ab1 peaks).

Gene Set Enrichment Analysis (GSEA) of the genes downregulated upon LOXL2 KD. The top 20 categories are shown, with the size of the points proportional to the adjusted P‐values and the distance from the center proportional to the gene ratio.
Schematic representation of BRD4L and BRD4S illustrating the Ab1 and Ab2 binding sites (top). Schematic representation of the ChIP‐seq strategy used to identify BRD4S preferentially bound sites (bottom).
Overlap of promoter target GSs of Ab1 and Ab2 identified with the MsigDB collections. The size of the points is proportional to the adjusted P‐values and the distance from the center is proportional to the gene ratio. The adjusted P‐values are calculated independently for each overlap comparison (Ab1 and Ab2).
RNA‐seq logFC for genes associated with the ChIP‐seq peaks of Ab2 in C which fall in promoter regions. The logFC of the subset of these genes which are DREAM targets are plotted in pink (top) and statistical significance is determined by permutation test (bottom).
BRD4 ChIP‐qPCR of DREAM target gene promoters in MDA‐MB‐231 cells infected with shControl (C), shBRD4 Long (BRD4L KD), and shBRD4 Short (BRD4S KD) isoforms. Data from qPCR were normalized to the input and represented as the fold‐change relative to the C condition, which was set as 1. Data are shown as the mean of three independent biological replicates. The standard deviation is shown as error bars. Significance was determined using a one‐way ANOVA multiple comparisons with Tukey's correction.
Genome Browser tracks of four different DREAM target genes containing the following information (from top to bottom): Ab1 ChIP‐seq profile, Ab2 ChIP‐seq profile either in C or LOXL2 KD conditions, and RNA‐seq signal in C and LOXL2 KD conditions.

Representative cell cycle profile of MDA‐MB‐231 cells infected either with C or LOXL2 KD. DNA content was analyzed by FACS following propidium iodide (PI) staining (left). The percentage of cells in each phase of the cell cycle was quantified using the FlowJo Software (right). Data are shown as the mean of three independent biological replicates. The standard deviation is shown as error bars. Significance was determined by unpaired Student's t‐test.
High‐throughput immunofluorescence of H3S10p mitotic marker in C or LOXL2 KD MDA‐MB‐231 cells. Mitotic cells (black dots) showed on average a higher H3S10p signal than the population median + 3S.D. Interphase cells are represented with gray dots. H3S10p intensity is represented as the normalized median. Significance was calculated using an unpaired Student's t‐test and is based on the mitotic index of the two populations.
Representative cell cycle profile of MDA‐MB‐231 cells treated either with DMSO or 40 μM of PXS for 96 h. DNA content was analyzed by FACS following propidium iodide (PI) staining (left). The percentage of cells in each phase of the cell cycle was quantified using the FlowJo Software (right). Data are shown as the mean of three independent biological replicates. The standard deviation is shown as error bars. Significance was determined by unpaired Student's t‐test.
MDA‐MB‐231 cells expressing SLBP‐mTurquoise2 and H1‐Maroon1 were treated with DMSO or 40 μM of PXS for 96 h. The percentage of mTurquoise2 nuclei in each well is shown, representing cells in G1‐S. The difference between the PXS and DMSO‐treated cells was significant (P < 0.005) under a linear model comparing the percentage of mTurquoise2‐positive cells, across time, in the two conditions. The estimated increase following treatment with PXS of the area‐under‐the‐curve (AUC) is 13.3 au. Significance was calculated by the Student's t‐test on AUC. Quantification was performed every 12 h. N = 200 cells/replicate, with six biological replicates.
Differential gene essentiality between high and low LOXL2‐expressing cell lines (CCLE) as calculated by analyzing the Achilles dataset. Cell lines with low LOXL2 expression are significantly more sensitive to the depletion of genes represented in the right part of the X‐axis as compared to cell lines with high LOXL2 expression, which are more sensitive to the depletion of genes represented in the left part of the X‐axis. N = 80 cell lines. Significance was determined using the Student's t‐test with BH multiple hypothesis correction. Significant threshold is based on adjusted P‐value < 0.05; black dots represent significant essentialities and pink dots represent different mediator subunits. The dot size is proportional to the adjusted P‐value of each gene.
Pulldown of endogenous Lin9, B‐Myb, or FOXM1 in MDA‐MB‐231 cells. Precipitates were analyzed by Western blot with the indicated antibodies. Irrelevant IgGs were used as a negative control; ns: non‐specific. Three biological replicates were performed.
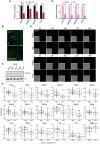
Real‐time quantitative PCR (qPCR) showing the changes in mRNA expression of four selected DREAM target genes (EZH2, HMGB2, AURKB, and PLK4) in C or LOXL2 KD MDA‐MB‐231 cells. Gene expression was normalized against an endogenous control (Pumilio homolog 1) and represented as the expression relative to the C condition, which was set as 1. Data are shown as the mean of three independent biological replicates. The standard deviation is shown as error bars. Significance was determined by unpaired Student's t‐test.
Representative images of H3S10P high‐throughput immunofluorescence in LOXL2 KD or C MDA‐MB‐231 cells. Scale bar, 100 μm.
Representative Western blot analysis of H3K4ox in MDA‐MB‐231 cells treated with DMSO or PXS for 96 h at the indicated concentrations. H3 was used as a loading control. Two biological replicates were performed.
qPCR showing the changes in mRNA expression of four selected DREAM target genes (EZH2, HMGB2, AURKB, and PLK4) in MDA‐MB‐231 cells treated with DMSO or PXS. Gene expression was normalized against an endogenous control (Pumilio homolog 1) and is represented as the expression relative to the DMSO condition, which was set as 1. Data are shown as the mean of three independent biological replicates. The standard deviation is shown as error bars. Significance was determined by unpaired Student's t‐test.
MDA‐MB‐231 cells expressing SLBP‐mTurquoise2 and H1‐Maroon1 treated with DMSO or PXS for 96 h. Representative images of the quantification in Fig 4D are shown. Images of big panels show SLBP‐mTurquoise2 (top) and H1‐Maroon1 (bottom); inset panels show their overlap with brightfield. Scale bar, 100 μm.
Gene‐specific differential gene essentiality between high and low LOXL2‐expressing cell lines (CCLE) as calculated by analyzing the Achilles' dataset. N = 80 cell lines. Significance was determined using the Student's t‐test with BH multiple hypothesis correction. The bottom and top fractions in the boxes represent the first and third quartiles, and the line, the median. Whiskers denote the interval between 1.5 times the interquartile range (IQR) and the median. Data beyond the end of the whiskers are plotted as outliers.
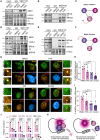
MED1 pulldown in MDA‐MB‐231 cells treated with DMSO or 40 μM of PXS for 96 h. Precipitates were analyzed by Western blot with the indicated antibodies. Irrelevant IgGs were used as a negative control; ns: non‐specific. Three biological replicates were performed.
BRD4 pulldown in MDA‐MB‐231 cells treated with DMSO or 40 μM of PXS for 96 h. Precipitates were analyzed by Western blot with the indicated antibodies. Irrelevant IgGs were used as a negative control; ns: non‐specific. Three biological replicates were performed.
Schematic representation of the interactions between BRD4S, LOXL2, and MED1 after LOXL2 inhibition with PXS.
MED1 pulldown in MDA‐MB‐231 cells treated with DMSO or 5 μM of JQ1 for 24 h. Precipitates were analyzed by Western blot with the indicated antibodies. Irrelevant IgGs were used as a negative control; ns: non‐specific. Three biological replicates were performed.
BRD4 pulldown in MDA‐MB‐231 cells treated with DMSO or 5 μM of JQ1 for 24 h. Precipitates were analyzed by Western blot with the indicated antibodies. Irrelevant IgGs were used as a negative control; ns: non‐specific. Three biological replicates were performed.
Schematic representation of the interactions between BRD4S, LOXL2, and MED1 after BRD4 inhibition with JQ1.
Representative images of BRD4 (green) and MED1 (red) high‐throughput immunofluorescence of MDA‐MB‐231 cells treated for 96 h with DMSO, 40 μM of PXS, 5 μM of JQ1 or the combination of both inhibitors (Combo). DAPI (blue) was used as a nuclear marker. Scale bar; 5 μm. Magnifications of representative foci are also depicted.
Quantification of the number of BRD4 foci from (G) corrected by the nucleus area. Results are normalized to DMSO. Four biological replicates were performed using at least 4,000 nuclei/replicate for the analysis. Data are shown as the mean of four independent biological replicates. The standard deviation is shown as error bars. Significance was calculated using a one‐way ANOVA multiple comparisons with Tukey's correction test.
Quantification of the percentage of MED1 overlapping with BRD4 occupied area from (G). Four biological replicates were performed using at least 4,000 nuclei/replicate for the analysis. Data are shown as the mean of four independent biological replicates. The standard deviation is shown as error bars. Significance was calculated using a one‐way ANOVA multiple comparisons with Tukey's correction test.
Real‐time quantitative PCR (qPCR) showing the changes in mRNA expression of four selected DREAM target genes (EZH2, HMGB2, AURKB, and PLK4) in MDA‐MB‐231 cells treated with DMSO, 40 μM of PXS, 5 μM of JQ1 or the combination of both inhibitors (Combo) for 96 h. Gene expression was normalized against an endogenous control (Pumilio homolog 1) and represented as the expression relative to the DMSO condition, which was set as 1. Data are shown as the mean of three independent biological replicates. The standard deviation is shown as error bars. Significance was calculated using a one‐way ANOVA multiple comparisons with Tukey's correction test.
Schematic representation of the proposed molecular mechanism.

Representative synergy matrixes showing cell viability measured with the MTT assay. The three TNBC cell lines used were treated with either PXS or JQ1 alone or with their combination at the indicated concentration for 96 h. SC indicates the synergy score for each cell line; a synergy score lower than 5 indicates the additive effect of the treatments, while a synergy score higher than 5 indicates synergism (Love et al, 2014). Three biological replicates were analyzed for each cell line.
Tumor volumes represented as fold change to day 1 (D1) from the MDA‐MB‐231 xenograft mice treated five times per week with 15 mg/kg JQ1 and/or 2 mg per pump PXS during 26 days. A minimum of six tumors per group (3 mice/both sides) are shown as average tumor volume and standard deviations are shown as error bars. The significance was determined at the endpoint (day 26) using a two‐way ANOVA multiple comparisons with Tukey's correction test (left). Images of the excised tumors at the end of the experiment (day 26) (right).
Quantification of the number of metastasis nodules per mouse lung section analyzed. Data are shown as the mean of at least 15 lung sections analyzed per group. The standard deviation is shown as error bars. Significance was calculated using a one‐way ANOVA multiple comparisons with Dunnett's correction test.
Tumor volumes represented as fold change to day 1 (D1) from PDX‐127 mice treated five times per week with 7.5 mg/kg JQ1 and/or 2 mg per pump PXS for 15 days. A minimum of six tumors per group (3 mice/both sides) are shown as average tumor volume. Standard deviations are shown as error bars. Significance was determined at the endpoint (day 15) using a two‐way ANOVA multiple comparisons with Tukey's correction test (left). Images of the excised tumors at the end of the experiment (day 15) (right).
Quantification of the number of H3S10p positive cells stained by immunohistochemistry in each of the excised tumors. A minimum of six tumors were analyzed per group and three different regions per tumor were quantified. The standard deviation is shown as error bars. Significance was calculated using a one‐way ANOVA multiple comparisons with Tukey's correction test.
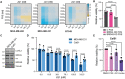
Representative matrixes showing the synergy score calculated with the cell viability data illustrated in Fig 6A.
Cell viability assay of MDA‐MB‐231 cells infected with shControl (C), shBRD4L (BRD4L KD), or shBRD4S (BRD4S KD) and treated with either DMSO or 20 μM of PXS for 96 h. Data were analyzed by DAPI count using the Operetta High Content Screening System and normalized to DMSO. Data are shown as the mean of three independent biological replicates. The standard deviation is shown as error bars. Significance was determined using a one‐way ANOVA multiple comparisons with Tukey's correction test.
Representative Western blot showing BRD4 and LOXL2 levels in MDA‐MB‐231 and Cal51 cell lines. Tubulin is shown as a loading control. Two biological replicates were performed.
Cell viability assay of MDA‐MB‐231 and Cal51 cells treated with the indicated concentrations of JQ1 for 96 h. Data were analyzed with MTT assay and normalized to DMSO. Data are shown as the mean of four independent biological replicates. The standard deviation is shown as error bars. Significance was determined using an unpaired Student's t‐test.
Cell viability assay of Cal51 cells treated with either DMSO, 40 μM of PXS, 312.5 nM of JQ1, or the combination of both (Combo) for 96 h. Data were analyzed with MTT assay and normalized to DMSO. Data are shown as the mean of three independent biological replicates. The standard deviation is shown as error bars. Significance was determined using one‐way ANOVA multiple comparisons with Dunn's multiple comparisons test.
Mouse body weight changes from the MDA‐MB‐231 xenograft mice treated as in Fig 6B at the endpoint (day 26). Weight changes are represented as the percentage with respect to day 1, and standard deviations are shown as error bars. Body weight changes of less than 10% are considered tolerable. A minimum of six tumors per group (with three mice per group, with one tumor on each side) are shown as average mouse body weight. Statistical analysis was performed using two‐way ANOVA multiple comparisons with Tukey's correction test for the whole experiment.
Representative Western blot showing LOXL2 and BRD4 protein levels of four different PDXs. Tubulin is shown as a loading control; ns: non‐specific. Two biological replicates were performed.
Tumor volumes represented as fold change to day 1 (D1) from PDX‐549 mice treated five times per week with 715 mg/kg JQ1 and/or 2 mg per pump PXS for 26 days. A minimum of six tumors per group (with 3 mice per group, with one tumor on each side) are shown as the average tumor volume. Standard deviations are shown as error bars. Significance was determined at the endpoint (day 26) using a two‐way ANOVA multiple comparisons with Tukey's correction test (graph). Images of the excised tumors at the end of the experiment (day 26) (pictures).
Mouse body weight changes from the PDX‐549 xenograft mice treated as in C at the endpoint (day 26). Weight changes are represented as the percentage with respect to day 1. Standard deviations are shown as error bars. Body weight changes of less than 10% are considered tolerable. A minimum of six tumors per group (with three mice per group, with one tumor on each side) are shown as average mouse body weight. Statistical analysis was performed using two‐way ANOVA multiple comparisons with Tukey's correction test.
Representative Western blot showing LOXL2 and BRD4 protein levels of 13 different PDXs. Tubulin is shown as a loading control; ns: non‐specific. Two biological replicates were performed.
Quantification of tumor doubling time measured as the day it reaches a volume of 2 as compared to day 1. For each PDX, the different number of tumors are shown (a minimum of three different tumors were analyzed). Data are shown as the mean of the replicates. Standard deviations are shown as error bars.
Mouse body weight changes from the PDX‐127 xenograft mice treated as in Fig 6D at the endpoint (day 15). Weight changes are represented as the percentage with respect to day 1. Standard deviations are shown as error bars. Body weight changes of less than 10% are considered tolerable. A minimum of six tumors per group (with three mice per group, with one tumor on each side) are shown as average mouse body weight. Statistical analysis was performed using two‐way ANOVA multiple comparisons with Tukey's correction.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- ERC-StG-852343/EC | European Research Council (ERC)
- INVES20036TIAN/Fundación Científica Asociación Española Contra el Cáncer (AECC)
- Fundación Francisco Cobos (FFC)
- LCF/BQ/DI19/11730061/'la Caixa' Foundation ('la Caixa')
- PID2019-110598GA-I00/MEC | Spanish National Plan for Scientific and Technical Research and Innovation (Plan Estatal de Investigación Científica y Técnica y de Innovación)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

