CSNK2 suppresses autophagy by activating FLN-NHL-containing TRIM proteins
- PMID: 37938186
- PMCID: PMC11135829
- DOI: 10.1080/15548627.2023.2281128
CSNK2 suppresses autophagy by activating FLN-NHL-containing TRIM proteins
Abstract
Macroautophagy/autophagy is a tightly regulated cellular process integral to homeostasis and innate immunity. As such, dysregulation of autophagy is associated with cancer, neurodegenerative disorders, and infectious diseases. While numerous factors that promote autophagy have been characterized, the key mechanisms that prevent excessive autophagy are less well understood. Here, we identify CSNK2/CK2 (casein kinase 2) as a negative regulator of autophagy. Pharmacological inhibition of CSNK2 activity or siRNA-mediated depletion of CSNK2 increased basal autophagic flux in cell lines and primary human lung cells. Vice versa, ectopic expression of CSNK2 reduced autophagic flux. Mechanistically, CSNK2 interacted with the FLN (filamin)-NHL domain-containing tripartite motif (TRIM) family members TRIM2, TRIM3 and TRIM71. Our data show that recruitment of CSNK2 to the C-terminal NHL domain of TRIM3 lead to its robust phosphorylation at serine 661 by CSNK2. A phosphorylation-defective mutant of TRIM3 was unable to reduce autophagosome numbers indicating that phosphorylation by CSNK2 is required for TRIM-mediated autophagy inhibition. All three TRIMs facilitated inactivation of the ULK1-BECN1 autophagy initiation complex by facilitating ULK1 serine 757 phosphorylation. Inhibition of CSNK2 promoted autophagy upon influenza A virus (IAV) and measles virus (MeV) infection. In line with this, targeting of CSNK2 or depletion of TRIM2, TRIM3 or TRIM71 enhanced autophagy-dependent restriction of IAV, MeV and human immunodeficiency virus 1 (HIV-1). Thus, our results identify the CSNK2-TRIM2, -TRIM3, -TRIM71 axis as a key regulatory pathway that limits autophagy. Targeting this axis may allow for therapeutic induction of autophagy against viral infections and in diseases associated with dysregulated autophagy.Abbreviation: ATG5: autophagy related 5; BafA1: bafilomycin A1; BECN1: beclin 1; CCD: coiled-coil domain; CSNK2/CK2: casein kinase 2; CSNK2A1: casein kinase 2 alpha 1; CSNK2A2: casein kinase 2 alpha 2; CSNK2B: casein kinase 2 beta; FLN: filamin; HeLa GL: HeLa cells stably expressing eGFP-LC3B; HIV-1: human immunodeficiency virus 1; IAV: influenza A virus; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3; MeV: measles virus; MTOR: mechanistic target of rapamycin kinase; RING: really interesting new gene; SQSTM1/p62: sequestosome 1; TRIM: tripartite motif; ULK1: unc-51 like autophagy activating kinase 1.
Keywords: Autophagy; ULK1; casein kinase; phosphorylation; tripartite motif proteins; virus.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures


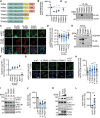

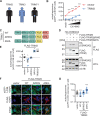

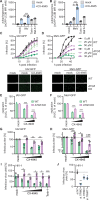
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
